|
Research Ideas and Outcomes :
Grant Proposal
|
|
Corresponding author: Jeannine Cavender-Bares (cavender@umn.edu)
Received: 02 Feb 2021 | Published: 05 Feb 2021
© 2021 Jeannine Cavender-Bares, Peter Reich, Philip Townsend, Arindam Banerjee, Ethan Butler, Ankur Desai, Amanda Gevens, Sarah Hobbie, Forest Isbell, Etienne Laliberté, José Eduardo Meireles, Holly Menninger, Ryan Pavlick, Jesús Pinto-Ledezma, Caitlin Potter, Meredith Schuman, Nathan Springer, Artur Stefanski, Pankaj Trivedi, Amy Trowbridge, Laura Williams, Charles Willis, Ya Yang
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Cavender-Bares J, Reich PB, Townsend PA, Banerjee A, Butler E, Desai A, Gevens A, Hobbie SE, Isbell F, Laliberté E, Meireles JE, Menninger H, Pavlick RP, Pinto-Ledezma J, Potter C, Schuman MC, Springer N, Stefanski A, Trivedi P, Trowbridge A, Williams L, Willis CG, Yang Y (2021) BII-Implementation: The causes and consequences of plant biodiversity across scales in a rapidly changing world. Research Ideas and Outcomes 7: e63850. https://doi.org/10.3897/rio.7.e63850
|
|
Abstract
The proposed Biology Integration Institute will bring together two major research institutions in the Upper Midwest—the University of Minnesota (UMN) and University of Wisconsin-Madison (UW)—to investigate the causes and consequences of plant biodiversity across scales in a rapidly changing world—from genes and molecules within cells and tissues to communities, ecosystems, landscapes and the biosphere. The Institute focuses on plant biodiversity, defined broadly to encompass the heterogeneity within life that occurs from the smallest to the largest biological scales. A premise of the Institute is that life is envisioned as occurring at different scales nested within several contrasting conceptions of biological hierarchies, defined by the separate but related fields of physiology, evolutionary biology and ecology. The Institute will emphasize the use of ‘spectral biology’—detection of biological properties based on the interaction of light energy with matter—and process-oriented predictive models to investigate the processes by which biological components at one scale give rise to emergent properties at higher scales. Through an iterative process that harnesses cutting edge technologies to observe a suite of carefully designed empirical systems—including the National Ecological Observatory Network (NEON) and some of the world’s longest running and state-of-the-art global change experiments—the Institute will advance biological understanding and theory of the causes and consequences of changes in biodiversity and at the interface of plant physiology, ecology and evolution.
INTELLECTUAL MERIT
The Institute brings together a diverse, gender-balanced and highly productive team with significant leadership experience that spans biological disciplines and career stages and is poised to integrate biology in new ways. Together, the team will harness the potential of spectral biology, experiments, observations and synthetic modeling in a manner never before possible to transform understanding of how variation within and among biological scales drives plant and ecosystem responses to global change over diurnal, seasonal and millennial time scales. In doing so, it will use and advance state-of-the-art theory. The institute team posits that the designed projects will unearth transformative understanding and biological rules at each of the various scales that will enable an unprecedented capacity to discern the linkages between physiological, ecological and evolutionary processes in relation to the multi-dimensional nature of biodiversity in this time of massive planetary change. A strength of the proposed Institute is that it leverages prior federal investments in research and formalizes partnerships with foreign institutions heavily invested in related biodiversity research. Most of the planned projects leverage existing research initiatives, infrastructure, working groups, experiments, training programs, and public outreach infrastructure, all of which are already highly synergistic and collaborative, and will bring together members of the overall research and training team.
BROADER IMPACTS
A central goal of the proposed Institute is to train the next generation of diverse integrative biologists. Post-doctoral, graduate student and undergraduate trainees, recruited from non-traditional and underrepresented groups, including through formal engagement with Native American communities, will receive a range of mentoring and training opportunities. Annual summer training workshops will be offered at UMN and UW as well as training experiences with the Global Change and Biodiversity Research Priority Program (URPP-GCB) at the University of Zurich (UZH) and through the Canadian Airborne Biodiversity Observatory (CABO). The Institute will engage diverse K-12 audiences, the general public and Native American communities through Market Science modules, Minute Earth videos, a museum exhibit and public engagement and educational activities through the Bell Museum of Natural History, the Cedar Creek Ecosystem Science Reserve (CCESR) and the Wisconsin Tribal Conservation Association.
Keywords
biodiversity, biological scale, global change, plant ecology and evolution, spectral biology
List of participants
David Coomes, University of Cambridge, UK, Science collaborator (2)
Lacey Hill-Kastern, Wisconsin Tribal Conservation Advisory Council (Education and outreach collaborator (1,3; III)
Eric Kruger, UW-Madison, USA, Science collaborator (1,4)
Richard Lankau, UW-Madison, USA, Science collaborator (1,3)
Rebecca Montgomery, UMN, USA, Science collaborator (4;VI)
Owen Petchey, UZH, Switzerland, Science collaborator (VII)
Matthew Ruark, UW-Madison, USA, Science collaborator (1,3)
Michael Schaepman, UZH, Switzerland,Science collaborator (1,2; VII)
Peter Thornton, Oak Ridge National Lab, USA, Science collaborator (4)
Scott Ollinger, University of Vermont, USA (ESAB)
Gabriela Schaepman-Strub, UZH, Switzerland (ESAB)
David Schimel, Jet Propulsion Laboratory, USA (ESAB)
Susan Ustin, University of California Davis, USA (ESAB)
Rationale and Justification
Biodiversity forms the basis of ecosystem function and the life support systems that promote human wellbeing. Advancing our understanding of Earth’s biodiversity and its response to global environmental change at scales from molecules to ecosystems is critical to societal capacity to adapt to and mitigate the loss of biodiversity in an era of rapid global change (
Decades of research on biodiversity and its relationship to ecosystem functions have revealed that variation in life on Earth matters for how ecosystems cycle elements, produce biomass, and respond to environmental change. Integration across biological scales is required to address fundamental questions that remain poorly understood including: 1) why and how life’s diversity matters at the largest scales—for biosphere function and dynamics critical to maintaining Earth’s life support systems—and 2) how and why variation at the smallest scales—genes and molecules within cells and tissues—influences processes at all other scales. The scientific advances required to tackle this set of problems have been hindered by the fragmentation of biology into specialized sub-disciplines that do not meaningfully connect these vastly different scales. Here we define the term ‘biodiversity’ not simply in its most common usage as species diversity but as a concept that also encompasses the variation in functional and evolutionary components within and among biological scales. As such our institute will address diversity at scales ranging from genomes to individuals and clades as well as from communities to ecosystems and the global biosphere.
One component of diversity is spectral variation, which results from chemical, anatomical, morphological, and architectural plant traits, that themselves may vary due to selection, evolutionary history, community composition and diversity, and environmental drivers. Spectral signatures can thus distinguish among different kinds of molecules in plants, as well as reveal variation across the range of scales we will study here: among leaves of individual plants, within and among species across the tree of life, and within and among plant communities, ecosystems, and landscapes across the global biosphere. ‘Spectral biology’ is thus a powerful and integrated means to capture biological variation—or biodiversity—across scales and to determine the causal factors that give rise to that variation (
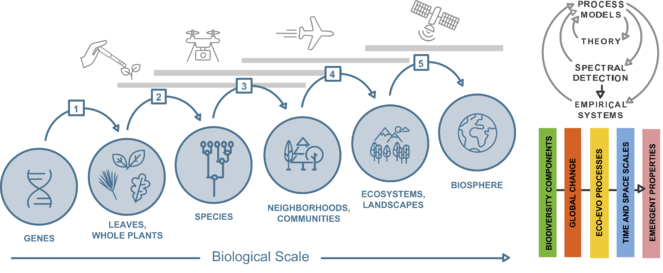
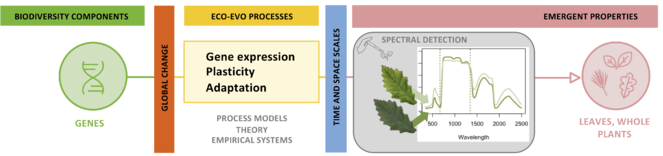
Conceptual framework (left), showing the five Themes of the Institute (1-5) as the transitions between biological scales. Each transition involves the interaction of biodiversity components (genes, leaves, species, communities, ecosystems) with environmental variation (global change forces) at different temporal and spatial scales resulting in emergent properties, which are the basis of biological variation at other scales. Three different concepts of biological scale (physiological, evolutionary, and ecological) are incorporated, as detailed in Fig.
Our primary mission is to uncover how the biological variation (i.e., diversity) and the processes that generate it at every scale influence processes and emergent properties at the next scale—ultimately to explain ecosystem and biosphere dynamics—and how changes in these global processes, in turn, drive biological variation at all other scales.
Conceptual Framework—Our overarching conceptual framework (Fig.
| Theme 1 | The genetic and environmental (i.e., GxE) drivers of trait variation at the leaf and whole plant scale linked by transcriptomic, metabolic and morphological variation |
| Theme 2 | How evolution generates the functional and spectral variation across the tree of life and its utility for biodiversity detection |
| Theme 3 | How functionally and spectrally distinct taxa, sampled across the tree of life, interact locally, leading to the assembly and dynamics of communities at multiple spatial scales, under current and future environmental conditions |
| Theme 4 | The consequences of biodiversity for ecosystem functioning and its response to global change |
| Theme 5 | How to improve parameterization of tissue- to ecosystem-scale properties at various spatial scales and advance land surface models that incorporate plant functional diversity |
In our conceptual framework (Fig.
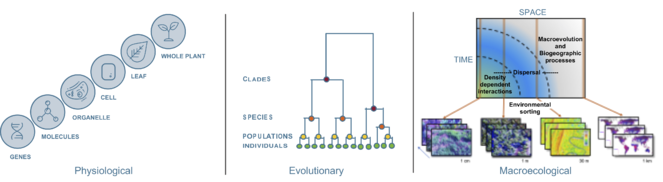
Three kinds of biological hierarchies that form the basis of biological integration and scaling. A) Physiological: Hierarchy of functional or metabolic units within a plant from genes, metabolites (small molecules) to organelles, cells, leaves to the whole plant. B) Evolutionary: Hierarchical organization of the tree of life from individuals to lineages. C) Ecological: Nested spatial scales showing that ecological and biogeographic processes—from density- and frequency-dependent neighborhood interactions to environmental sorting, dispersal and biogeographic processes—that drive the distribution and diversity of life and shift with increasing spatial and temporal scales.
We consider emergent properties as observable phenomena that represent aggregate functions of biological systems at each scale of inquiry that result from biological components and their interactions with each other and the environment at the scale below. Emergent properties thus arise from various interactive processes of system components but are more than the sum of the quantifiable parts (
Projects both within and spanning these Themes are designed to elucidate the biological components and underlying processes that give rise to the emergent properties of biodiversity—including the emergent spectra—at multiple scales. Projects include common garden studies of model species in which we identify genetic interactions with the environment—including climate, pests, pathogens, and the soil microbiome—and modeling of evolutionary processes within model clades and across the plant tree of life that give rise to functional and spectral trait variation among species (Themes 1 and 2). They also include unparalleled global change experiments (Themes 3 and 4) and NEON-based biodiversity observations (Themes 4 and 5). At the community and ecosystem levels, we experimentally test how species interactions and dispersal lead to biodiversity (taxonomic, phylogenetic, functional, spectral) at multiple spatial scales, with consequences for carbon, water and nutrient cycles under current and future environmental conditions. Finally, at the planetary scale, spectral biology—in concert with machine learning and simulation modeling—provides the potential to represent all of these finer-scale processes within the context of biosphere-atmosphere interactions using models that upscale ecosystem emergent properties and functions to the globe (Theme 5). Collectively, the research Themes and projects within and across them provide a critical platform for training the next generation of integrative biologists. Through graduate student and postdoctoral training, summer training workshops and winter symposia, development of public exhibits and videos tailored to K-12 audiences and the public, and opportunities for international exchange with related programs in Switzerland and Canada, these collective projects provide the opportunity for education and public outreach about life’s variation. In doing so, we will engage diverse audiences in new understanding of the connectedness of biodiversity from genetic variation to the tree of life and how its losses affect our human habitat and life support systems. Our highly collaborative and diverse team brings together expertise and skill sets that will enable us to tackle these challenging themes.
Organization and Management Design
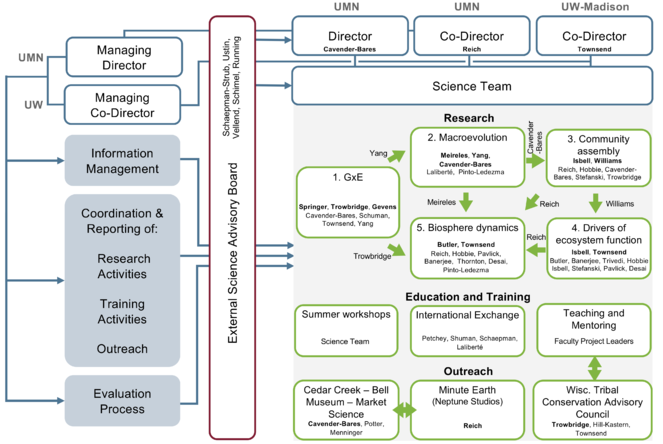
The Institute will have a Director and two Co-Directors, responsible for managing the research activities within and across all project Themes (Fig.
Schematic of the Management Plan, showing management structure and responsibilities of personnel in relation to Research, Education and Training, and Outreach components of the Institute. The Directors will oversee all research, education and training, and outreach activities. Project Leader(s) (shown in bold) will be designated to oversee each of the five thematic research areas along with Project Leaders of specific projects within the thematic area (not in bold). Project Leaders who will oversee bridging and integration activities are shown next to green arrows linking the thematic areas. One Project Leader (shown in bold) will oversee each of the three major outreach activities in collaboration with other Project Leaders and collaborators. All Project Leaders will collaborate in education and training activities. A Managing Director and Co-Managing Director, along with an information management team, will ensure data are managed properly, and will coordinate communication; research, education and training, and outreach activities; the evaluation of those activities; and reporting functions. The External Science Advisory Board contributes to evaluation and provides feedback on research, education and training, as well as outreach.
Leadership and Team
Our highly productive and collaborative team spans biological sub-disciplines within evolution and physiological and ecosystem ecology with expertise in genetics, '-omics' approaches, bioinformatics, global change biology, spectroscopy, and modeling. We conduct predictive modeling in all of these domains. Collectively our team has forged innovative and integrative research in plant biology that has transformed our understanding of plant hierarchies, the interconnection of ecological and evolutionary processes in community and global change ecology and the role of spectral biology in bridging biological scales to understand patterns and consequences of biodiversity. Director Cavender-Bares is an integrative physiological and evolutionary ecologist and a leader in harnessing phylogenetic and spectroscopic methods in ecology. She previously led a NASA/NSF Dimensions of Biodiversity grant with Townsend, Hobbie, Meireles, Reich, and Williams to use spectral biology to link genetic, phylogenetic, and functional dimensions of biodiversity above and belowground across scales (
The External Scientific Advisory Board (ESAB), composed of eminent scientists, will meet annually and provide strategic guidance, vision, and accountability for the Institute (Fig.
Risk Management Strategies
The Directors and Project Leaders have a strong history of collaboration, inclusive authorship, and promoting early career scientists to take leadership on publications. If a scientific disagreement arises, we will attempt to resolve it through collegial discussion or seeking guidance from colleagues. In the unlikely event mediation is needed to resolve a dispute, Dr. David Greenstein, Associate Dean for Research in the College of Biological Sciences at UMN will provide mediation. If a project ends (see Feedback and Evaluation), the Directors will jointly determine how best to reallocate resources to optimize meeting the overall Institute goals. If any dispute arises that cannot be resolved by mutual agreement after meetings and mediation, it will be finally settled under the Rules of Conciliation and Arbitration of the American Arbitration Association by one arbitrator appointed in accordance with the Rules.
Feedback and Evaluation
The Institute will monitor projects through a two-pronged approach. First, projects will be monitored throughout the year by the Managing Directors for the projects’ progress towards completing milestones and meeting objectives as delineated in project work plans. The Directors and Managing Directors will have regular (minimum quarterly) check-ins with Project Leaders to assess progress. When milestones are not being met, the Directors and Managing Directors will determine the adjustments required to get the project back on track. In some instances, project delays may be due to conditions beyond the Project Leader’s control. In these circumstances, the delay will be documented and necessary adjustments made. If the delay were to result in an inability to complete the project, a meeting would be held among the Institute Directors and the Project Leader to determine if the project should end. Second, a formal annual assessment process will be led by the Managing Directors with the ESAB and experts recruited specifically for individual projects serving as a Research, Education and Outreach Evaluation and Advisory Panel (REAP), coincident with an annual symposium (see below). An approach similar to the mid-term reviews of NSF LTER projects will be adopted. Each panelist will review all projects in their assigned area. The review process will include a two-step process: 1) REAP members will review and score projects independently and 2) the Institute will convene REAP members via conference calls to discuss and provide final recommendations for project advancement and adjustments. REAP recommendations will be reviewed by Institute Directors. Evaluation criteria will explicitly include biology integration advancement, integrative training advancement and transdisciplinary collaboration. In addition, the research projects will be reviewed and scored for milestone completion and deliverables achieved (or on target). Prior to the annual REAP assessment, Education and training efforts will be assessed by a faculty team at UMN and UW, led by Willis as part of the highly acclaimed Biology Teaching and Learning Department at UMN. The process will include an assessment of pre-defined learning outcomes, learning success indicators, and students’ sense of inclusivity and confidence pre- and post-participation. Post-doctoral evaluation is addressed in the Post-doctoral Management Plan. Outreach evaluation processes for the Bell Museum, Cedar Creek and Market Science activities will be led by Cavender-Bares in collaboration with Menninger and Potter and evaluation of Minute Earth activities will be led by Reich in collaboration with Neptune Studios; both have well-established evaluation procedures in place. Trowbridge will lead evaluation processes for engagement and training activities with the Wisconsin Tribal Conservation Association. These evaluation procedures will be reviewed by the REAP and the ESAB. Education/training and outreach efforts will be adjusted based on review and assessment.
Evaluation Results
For multi-year projects, reviewers will use the annual review scores and assess the project goals, deliverables, methods, and feasibility for the upcoming year. If the project scores high and reviewers deem the project plan feasible, the project will continue. The evaluation plan will serve to assess the overall success of the Institute and inform and adjust Institute priorities and investments. It is intended to foster program transparency, connect multiple evaluation activities, and provide a comprehensive document that articulates the plan components.
Annual Symposium
Every winter, the Managing Directors and Project Leaders will organize a symposium to be held at UMN or UW in alternating years in which all postdocs and students will present research results. The ESAB will attend these events in person, if possible, or via video when necessary. The Managing Directors and Project Leaders will prepare an annual report documenting results within each project Theme and include abstracts of each subproject. These will be presented orally or with posters at the symposium. One day of the symposium will be open to the public, and an ESAB member may be invited to speak. The ESAB will provide an annual evaluation of the Institute’s progress and offer feedback for improvement. ESAB members were chosen based on their experience in managing projects of similar scope and scale.
INTELLECTUAL MERIT
Research Activities
Conceptual Background and Current Evidence
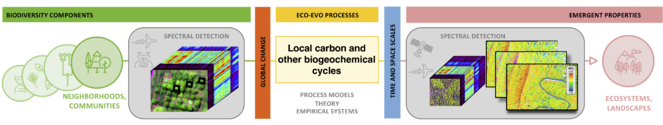
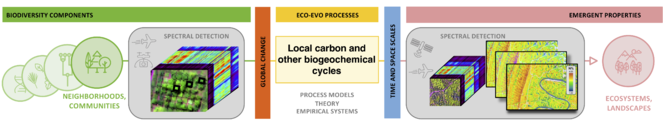
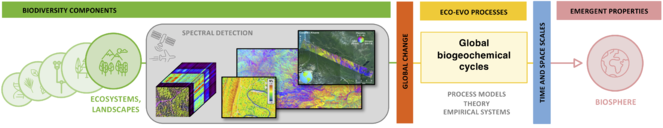
The problem. Biology has increasingly fragmented into subdisciplines, posing challenges to uncovering unifying principles and integrating understanding across biological scales. This lack of integration is particularly problematic for understanding evolutionary sources of biological diversity, its local and global responses to global change threats, and the consequences of these changes for ecosystem functions and biosphere dynamics. Such understanding is critical for managing threats to life support systems. We propose to advance integration of biology by examining plant biological variation in response to environmental variation across a range of scales, using a common conceptual framework and set of integrative tools. Specifically, spectral observations at each scale will serve as information-rich inputs to predictive models developed from theoretical understanding of biological processes to yield unifying principles that are critical to advancing knowledge of biodiversity change and its impacts. We view biodiversity as multi-faceted, encompassing biological heterogeneity at levels from genes to whole plants, species, communities, ecosystems, landscapes and the globe. We investigate the processes by which life scales across hierarchies, space, and time and ask how biodiversity at one hierarchical level interacts with environmental variation to influence emergent properties at the next higher scale (Fig.
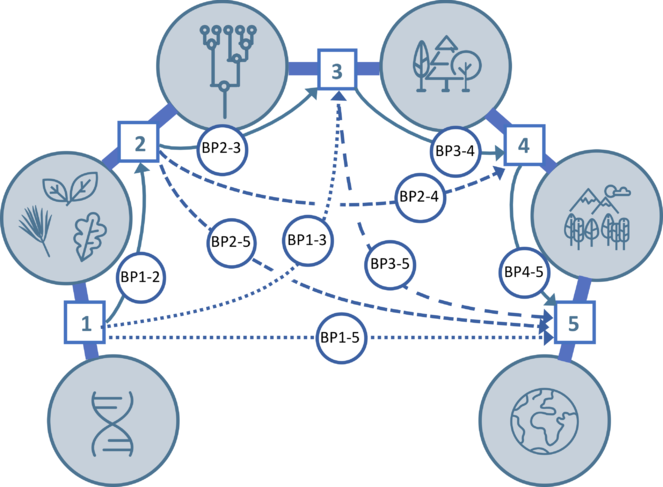
The five project Themes—1. Genetic and environmental drivers of trait variation, 2. Evolution of variation in traits across the tree of life, 3. Community assembly dynamics, 4. Biodiversity consequences for ecosystems, 5. Advancing tissue- to ecosystem-scale modeling of Earth’s land surface—and the bridging projects (BP) that integrate them, indicated by connecting arrows.
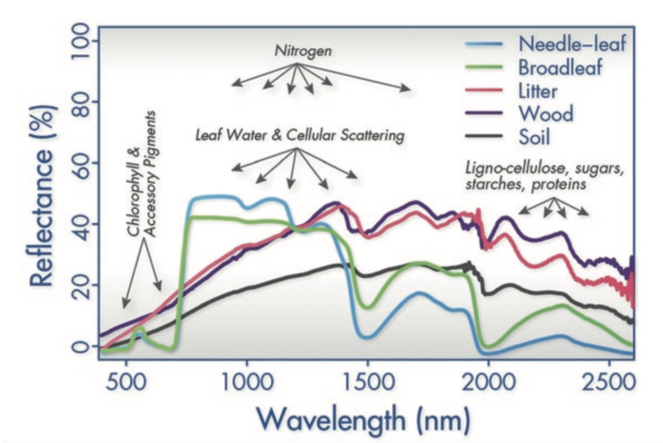
The spectral approach. We use 'spectral biology' to detect diversity at each biological scale. Spectral information will be coupled with genetic, physiological, community, and ecosystem data and integrated using process-based models to predict processes and patterns across scales. 'Spectral biology' harnesses information from plant electromagnetic spectra—the patterns of light absorbed, transmitted, and reflected at different wavelengths—that are aggregate indicators of the chemistry, morphology, and physiology of leaves, canopies, communities, and ecosystems (Fig.
Electromagnetic spectra of plants and ecosystem components with an indication of the underlying biochemistry from (
Building predictive models for associating spectral signals with functional diversity within and among species and ecosystems. Spectral biology is powerful because it provides concurrent insights into multiple factors that shape plant and ecosystem function. The same spectra that can predict chemical and morphological features in plant leaves and canopies (
Scaling from leaf-level functions and community interactions to ecosystem properties. An extensive literature on “biodiversity and ecosystem function” (BEF) (
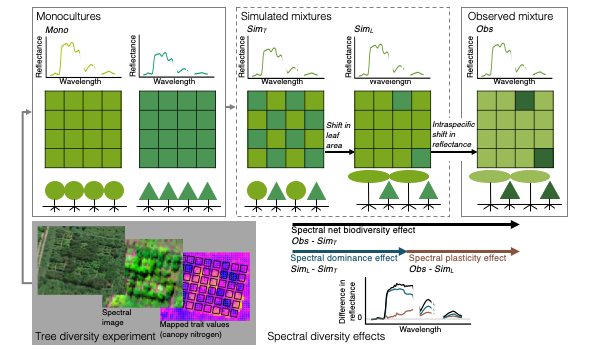
Remote spectroscopic imaging of a tree diversity experiment allows us to assess diversity effects on spectral reflectance, functionally important canopy traits (e.g. %N) and ecosystem functions (e.g. productivity) as well as underlying ecological processes. By simulating the spectra of mixed-species stands from monocultures, we can calculate the spectral net biodiversity effect (following a field-based approach) and partition novel, ecologically informative contributions to this net effect, such as spectral dominance and spectral plasticity effects. In this hypothetical example, two species (circles, triangles) are depicted in monoculture, simulated mixtures, and observed mixture. Each assemblage is illustrated in cross-section, from overhead (grid represents pixels), and as a mean reflectance spectrum.
Linking biodiversity-ecosystem function relationships to global biosphere dynamics through Earth system models (ESMs). ESMs, currently our best means of predicting biosphere responses to global change, dramatically oversimplify the functional diversity of vegetation by lumping all vegetation into a small number (≈ 4-14) of plant functional types (PFTs). Moreover, each PFT (e.g., temperate broad-leaved deciduous or tropical evergreen rainforest trees) is prescribed a single value (the mean of available measurements) for critical traits. Consequently, most models treat enormous areas of biodiverse vegetation as functionally and chemically identical. In reality, there is considerable heterogeneity of traits within and among ecosystems across Earth (
Core Themes and Synergistic Projects
We outline a set of synergistic collaborative projects within and across five Themes (Figs
Theme 1. The genetic and environmental drivers of trait variation at the leaf and whole plant scale linked by transcriptomic, metabolic, and morphological variation (Fig.
We will examine the mechanistic basis for spectral and functional variation within a species, scaling from the variation in genes (left), transcriptomes, and metabolites to the variation that emerges in leaves, canopies, and genetically distinct individuals exposed to contrasting environments (right).
Spectroscopy has the potential to document genetic variation (
1.1 Abiotic global change experiments in controlled environments. In growth chambers, we will assess GxE interactions on spectral traits by growing seedlings of at least ten different genetic backgrounds (i.e., genetically identical individuals for crops, or siblings within a seed family for trees). Seedlings will be exposed to different environmental stresses including control, cold, heat, and salinity. Also, replicated plantings of corn and oak from each of 10 genetic backgrounds will be placed within the rings of the BioCON experiment at the Cedar Creek LTER to impose the same CO2 treatments as in Theme 3.
1.2 Common gardens across climatic and biotic gradients. We will explore traits connecting genetic variation and GxE interactions to spectral variation by leveraging four existing sets of studies: (1) Pinus ponderosa genetic trials (200 half sib families) in five sites across MT and ID and a seed orchard in WA with 28 genotypes; (2) Georeferenced beech (Fagus sylvatica)-dominated forests, extant biodiversity plantations that include F. sylvatica, a common garden experiment with 100 seed families in Zurich in which climatic factors (water, soil, herbivory) can be manipulated, and 5 common gardens with 20 seed families across a Swiss climatic gradient (these experiments, campaigns, and sample/data analysis are fully supported by the UZH and URPP-GCB); (3) The Adaptation to Climate reciprocal transplant Experiment (ACE) that has 60 seed families of bur oak (Quercus macrocarpa) growing in a nursery to be planted in common garden sites across a climatic gradient in MN, IL, and OK; and (4) 9 potato trials established in 9 states with 10 genetic backgrounds (USDA-NIFA funded). We will identify plant responses (physiology, yield, quality) to soil microbiome diversity, pathogen inoculum, and pathogen suppression, leveraging data on microbial community composition via sequencing of prokaryotic 16S-V4 and fungal ITS2 rRNA gene regions. For each of these sites we will leverage micrometereological data and collect leaf and whole canopy spectra and species-specific phenotypic data to associate with metabolomic and transcriptomic profiles.
1.3 Field manipulation experiment. We will further leverage a drought experiment at the Sevilleta LTER near Socorro, NM focused on pine (Pinus edulis) that uses rainout shelters to divert 45, 75, and 90% of ambient precipitation away from trees. We will access micrometeorology, soil moisture, and sap flow data, as well as PhenoCam photos, chemical defenses, non-structural carbohydrates, physiological measurements, and insect community composition. Leaf and whole canopy spectral traits will be collected immediately before tissue samples are collected for transcriptome and metabolomic profiling.
Methods for linking transcriptomics, metabolomics, and spectral reflectance. For all Theme 1 projects, we will generate RNAseq based transcriptomes and metabolomic data using established protocols. For each RNAseq sample, we will generate ~20M RNAseq read pairs on an Illumina platform, to determine transcript abundance for each gene. For metabolomic profiling, pigments, natural products, and small signaling molecules (e.g., defense compounds, hormones) will be analyzed using liquid chromatography (HPLC-UV) at UMN and mass spectrometry-based profiling (UHPLC-MS/MS profiling) (
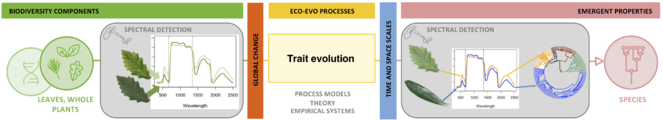
Theme 2. How evolution generates the functional and spectral variation across the tree of life and its utility for biodiversity detection (Fig.
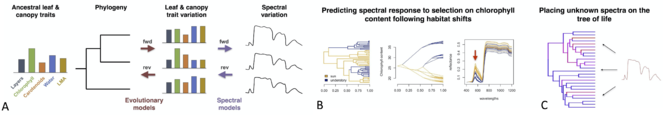
Functional and spectral variation among species and lineages is critical for remotely detecting biodiversity in plant communities. Although such variation arises from macroevolutionary processes that involve convergence, divergence, and constraints in plant structure and chemistry, the effects of evolution on plant spectra are poorly understood. By coupling macroevolutionary models and physical models of leaf spectral properties (Fig.
Integration of trait evolution and leaf spectral models enables estimation of evolutionary parameters from spectra and simulation of leaf spectra along a phylogeny. Ancestral leaf attributes evolve along a phylogeny, generating the current leaf attributes that underlie spectra. From the evolved leaf attributes, radiative transfer models estimate spectra that carry the signature of the phylogeny (A). This framework allows us to infer the evolutionary dynamics of spectra and to predict spectral responses to evolutionary pressures (B) as well as to compute the probability that an unknown spectrum falls anywhere on the tree of life (C).
2.1. Macroevolutionary modeling of plant spectral and other functional traits
2.1.1. Sampling across the tree of life. We will examine the origins of diversity in functional and spectral traits observed across the plant tree of life using multivariate Ornstein-Uhlenbeck models to estimate evolutionary rates, constraints (or directional evolution), lability, and coordination in plant traits (
2.1.2. Comprehensive sampling within two model clades. Using comprehensive species sampling in two broadly distributed lineages, we will explore the mechanistic links between genetic, metabolic, and spectral variation and long-term evolution. We will measure leaf-level spectra from living collections for ca. 300 (of 450 known) Quercus species and ca. 60 (of 120 known) Pinus species from arboreta in the US, France, England, and Mexico. Mature leaves will be collected and flash-frozen following established protocols for RNA-seq
2.2. Biodiversity detection and modeling. We will spectrally identify species, lineages, and community types through remote sensing across scale (
Theme 3. How functionally and spectrally distinct taxa, sampled across the tree of life, interact locally, leading to the assembly and dynamics of communities at multiple spatial scales, under current and future environmental conditions (Fig.
Evolutionary processes involved in generating the variation within and among species across the tree of life (left) provide the raw materials—taxa and their functional traits—for the processes (growth, survival, dispersal) by which individuals interact, leading to coexistence or to competitive exclusion and dominance, in local communities and metacommunities (right).
The composition and diversity of ecological communities emerge from species interactions within and among local communities. To address Theme 3, we will leverage (1) long-term forest and grassland manipulations of global change factors and (2) a new dispersal experiment in grasslands. These experiments allow the assessment of how trait variation, individual species performance, and plant-plant interactions drive community assembly processes and associated diversity. As platforms for this research, we will use ongoing long-term open-air field experiments in Minnesota (9 to 23 years-old) in which CO2, temperature, rainfall, N deposition, and/or plant biodiversity are manipulated [B4WarmED, IDENT (IDENT-Cloquet, FAB-1 and FAB-2), BioCON, TeRaCON] (
3.1 Community Assembly (BioCON/TeRaCON, RESCUE, B4WarmED and IDENT)
3.1.1. Grassland global change experiment. In the nested multi-factor (temperature, rainfall, CO2, N and diversity) BioCON/TeRaCON experiments, we will test (1) whether outcomes of species interactions in mixtures under global change can be predicted from traits of individual species observed in monocultures, and (2) how global changes alter local coexistence, favoring some species over others. For (1) we will assess whether species that drive resources (nutrients, light, water) to the lowest levels in monoculture outcompete other species in mixtures, as predicted by resource competition theory (
3.1.2. Metacommunity grassland experiment. In RESCUE we will manipulate species dispersal, a metacommunity process (
3.1.3. Forest diversity and global change experiments. In the B4WarmED (
3.2 Neighborhood carbon cycle (B4WarmED and IDENT). In a subset of Theme 3 studies, we will use leaf-scale daily and seasonal physiological relations (
Theme 4. The consequences of biodiversity for ecosystem functioning and its response to global change (Fig.
The integration of evolutionary and community assembly processes (left) result in emergent properties of communities (e.g. leaf area index, canopy %N) that drive ecosystem processes (right) and their responses to global change. To address Theme 4, we will leverage the same set of long-term field experiments described above (Theme 3), complementing existing data with additional modeling and measurements.
4.1 Community Scale Traits and Biogeochemical Cycles. In all seven global change experiments (Theme 3), we will (1) test whether the mean and variance of community-scale traits explain system-scale C and N cycling and its responses to global change factors (
These experiments (1) represent some of the longest-running, most complex, well-replicated, multi-factor, and ecologically realistic global change experiments of their kind, (2) contain existing taxon-specific trait data (including spectral data) and will be platforms for generating new data of this type during the BII, and (3) provide a rich data infrastructure, as a number of ecosystem properties and processes (e.g., biomass production, soil C flux, soil N cycling, plant and soil C and N pools) have been measured in every plot; allowing benchmarking of process models that use emergent properties (including spectrally derived ones) to predict ecosystem scale responses to global change factors (
4.2 Plant-soil Interactions. Plant-soil feedbacks are interactions among plants, soil organisms, and abiotic conditions that influence plant performance, diversity, and community structure, ultimately driving ecosystem processes (
Theme 5. How to improve parameterization of tissue-to ecosystem-scale properties at various spatial scales and advance land surface models that incorporate plant functional diversity (Fig.
Building on ecosystem scale estimates of plant functional diversity (left), we will test the effect of diversity on ecosystem functioning across large spatial extents, including by constructing global maps of functional diversity and incorporating processes described in Themes 3 and 4 into global scale models to simulate the influence of diversity on biosphere dynamics (right).
5.1 Functional diversity and ecosystem function in NEON. Using hyperspectral imagery, we will quantify functional diversity spatially to develop and test BEF relationships, to inform incorporation of biodiversity into global land surface models. First, we will leverage flux tower and hyperspectral imagery from the NEON to identify relationship between functional diversity (FD) and gross primary production (GPP) at broad scales (see Research Resources section). We will calculate FD following
5.2 Diversity and function at the landscape scale. Similar approaches can be applied to test diversity-function relationships (
5.3 Diversity and function at the global scale. At the global scale, incorporation of BEF relationships into models of current vegetation function will require remote sensing for both the characterization of functional diversity and ‘ecosystem function’ (as opposed to flux towers, as above), and into prognostic models of future vegetation will require modeling (rather than measuring) future trait diversity. For the former, we will address the potential to measure BEF relationships entirely using remotely sensed data, leveraging numerous datasets (
For simulating diversity effects under global change, both traits and responses must be modeled. Spectral data will be incorporated into terrestrial biosphere models through the generation of global gridded maps of trait diversity and their variation with environmental change, leveraging hyperspectral-derived trait maps and ground-based data. Spectral data will be used in conjunction with a global database of in situ trait measurements (TRY;
Our site-based simulations provide the basis for incorporating trait distribution maps and novel processes into simulations of the global carbon cycle and will be evaluated against: 1) the ILAMB model evaluation metrics (
The analyses proposed here will pave the way for much broader analyses that will be possible once global satellite imaging spectroscopy data for mapping functional diversity become available in the mid- to late-2020s from the planned NASA Surface Biology and Geology (SBG) and ESA Copernicus Hyperspectral Imaging Mission (CHIME) missions. This effort represents an unprecedented integration of ground-based data, remote-sensing data, spectral ecology, trait-based ecology, machine learning, and biodiversity-ecosystem function science and expertise, which together will enable large advances in understanding and quantifying biosphere-atmosphere interactions influencing the global carbon cycle.
These analyses also provide an empirical basis for incorporating functional diversity into terrestrial biosphere models, with spectral biology providing the necessary foundation.
Bridging Projects: Integration across Themes. Bridging research (BP1-2, BP1-3, etc., Fig.
BP1-2, 1-3. Intensive species-specific studies (Theme 1) will improve understanding of how functions of species have evolved in association with shifts into habitats and ecological niches that vary in precipitation and temperature. These insights will help us develop generalizations about trait evolution (including spectral traits) across the tree of life (Theme 2) and about species’ responses—including those that shape interactions among plants and drive community change—to global change factors (Theme 3).
BP2-3, 2-4. The macro-evolutionary processes involved in generating variation among species (Theme 2) provide the raw materials—taxa and their functional traits—for the processes (establishment, growth, survival, dispersal) by which individuals interact, leading to coexistence or competitive exclusion, in local communities and metacommunities (Theme 3) and to the aggregate community traits that drive ecosystem processes (Theme 4). We will test whether spectral signatures derived from the tree of life project (Theme 2), and extrapolated to model systems (Themes 3 and 4), predict traits that explain species competitive outcomes (Theme 3) and ecosystem function (Theme 4)—and how spectra and competitive outcomes vary with global change context. We will compare multiple approaches for predicting outcomes of neighborhood interactions and resulting ecosystem functions (
BP3-4. The outcomes of neighborhood interactions and community assembly processes (Theme 3) will result in the functional composition that drives ecosystem outcomes (Theme 4). Here we will address (1) whether the traits that lead to success for species in competition under varying global change contexts predict the magnitude and variation of ecosystem process rates in each context and (2) whether common spectral signatures predict shared responses at species, community, and ecosystem scales.
BP1-5. Physiological and spectral relations with climate of origin for bur oak and ponderosa pine (Theme 1) will be incorporated into parameterization of their respective clades at appropriate regional scales in land surface modeling (E3SM or CABLE) (Theme 5). Resulting model output (e.g. GPP) will be evaluated against US (FIA) and Canadian (CFS) inventory data, and/or flux tower data (e.g.,
BP2-5. Improved quantification of how spectral signatures map onto the tree of life (Theme 2) will be employed as part of new models of global traits developed from machine learning applied to combinations of existing data bases (TRY) and global satellite spectral data, a key foundation of Theme 5.
BP3-5. Advances in understanding variation in species performance under experimental environmental change, and how variation in spectral signatures and other emergent properties relate to such performance (Theme 3), will be incorporated into process models (G’Day, STANDLEAP, E3SM) that operate at ecosystem to global scales (Theme 5). Model parameters will be updated using both direct (photosynthesis, growth) and spectral scaling relationships (Theme 3). Similarly, the C cycle-environmental response logic (e.g. temperature response of photosynthesis or respiration) required to integrate and scale from leaf to ecosystem will be improved to better incorporate heterogeneity across taxa and environments (
BP4-5. Conceptual and quantitative advances in understanding how biodiversity influences the magnitude and stability of ecosystem processes, such as plant productivity (
Research Resources
Our institute makes extensive use of existing NSF research investments and resources, including NEON, LTER, as well as resources funded by USDA NIFA, NASA, and the University of Zurich (UZH).
Theme 1. Common Garden Resources with permission to fly drones. Pine common gardens. Genetic trials of Pinus ponderosa are maintained by Inland Empire Tree Improvement Cooperative (IETIC), with the same 200 half-sibs (5 replicates) across sites that differ in micrometeorology and disturbance regimes (e.g. drought and bark beetles). The Pullman Ponderosa Pine Seed Orchard (PPSO) was established in May 2007; 28 known genotypes were grafted to seedling rootstocks and planted with 10-16 replicates of each. We also leverage a recently established large-scale manipulative drought experiment (NSF-funded) at the Sevilleta LTER, NM that includes Pinus edulis and consists of five 40 x 40 m plots with control, 45%, 75%, and 90% reduction of rainfall, and a 10-yr legacy plot with 45% reduction. Micrometeorology, soil moisture, tree water content, sap flow, and PhenoCam photos (half-hourly) and monthly collections of needle and twig samples for terpene analysis and non-structural carbohydrate (NSC) analysis, water potential, tree canopy health, and Lindgren funnel collections of the arthropod community in each plot; bole xylem, phloem and roots samples for terpene analysis and non-structural carbohydrates and physiological measurements, and bole and branch volatile organic compound emissions are available. Potato common gardens: Field trials of potato varieties have been established in 9 states (CO, ID, ME, MI, MN, ND, OR, WA, and WI), with 12 m x 4 m plots and 6 agronomic treatments each replicated five times, to be sampled annually through 2024. Data available to us includes microbial community composition in every soil sample via sequencing of prokaryotic 16S-V4 and fungal ITS2 rRNA gene regions, as well as crop health, yield, and quality data. The project is funded by USDA-NIFA. Oak common gardens: The Adaptation to Climate (ACE) experiment is in progress with one-year seedlings of sixty seed families of Quercus macrocarpa to be planted in common garden sites across a climatic gradient in MN, IL and OK. Ten replicates of each of the 60 seed families from the three sources will be planted 1 m apart in a randomized block design at CCSER, Morton Arboretum and U OK Kessler Field Station, supported by CCESR NSF-LTER and UMN. Beech common gardens: Fagus sylvatica common garden experiments are currently being established with 100 seed families in one garden at the UZH, and 20 seed families in gardens across a climatic gradient in Switzerland. Maize inbred lines are currently in the Springer lab germplasm collection and are publicly available through the USDA North Central Plant Introduction Station.
Theme 2. Leaf spectra from the tree of life and the oak model clade. A database of 560 species (and replicates of species) has been assembled through NSF-NIMBioS (
Themes 3 and 4. Tree diversity experiments: IDENT - Cloquet located in northern Minnesota in which species richness and functional diversity have been independently manipulated; 48 different species assemblages (of 1, 2 and 6 species) were planted in replicated plots. The location of all trees (9408) is known and annually measured since planting (11 census points). An IDENT Canopy Carbon Model estimates species and stand-level carbon assimilation at hourly time steps across the experiment using incident radiation, leaf area, photosynthetic light response curves, and foliar phenology to estimate total C assimilation. The IDENT - Forest and Biodiversity (FAB1) experiment at CCESR has a similar design as IDENT-Cloquet. A larger FAB2 experiment with 10x10 m and 20x20 m plots manipulates taxonomic and phylogenetic diversity of trees from 1, 2,4,6 and 12 species mixtures that vary in functional and phylogenetic diversity; these have been supported by NSF-LTER, NSF/NASA, and UMN. B4Warmed located at two sites in northern Minnesota (Cloquet, Ely) is the world’s longest running open-air active warming experiment (2020 will be year 12), and one of only two (the other TeRaCON) that control temperature elevations both aboveground and belowground. It is well replicated (72 plots, >10,000 tree seedlings) and includes factorial combinations of temperature, rainfall manipulation, sites and habitat type (open, understory). It has been supported by the US DOE and Minnesota’s LCCMR.
BioCON/TeRaCON at CCESR, is the world’s longest running free-air CO2 enrichment experiment, and includes 16 factorial combinations of CO2, N supply, and plant biodiversity in a well replicated (371 plots) 22-year old grassland experiment. TeRaCON is a sub-experiment using 48 of the BioCON plots, which deploys 16 combinations of warming, rainfall manipulation, CO2, and N supply; it is one of only several such multi-factor long-term global change experiments (2020 will be year 9) and has received considerable NSF support through the LTER, LTREB, MRI, Ecosystems and Biocomplexity programs.
The RESCUE experiment (to be established in 2020 as part of NSF CAREER award to Isbell) will fully cross a habitat destruction treatment (loss of 0%, 50%, 95%, or 99% of local habitat area) with a seed addition treatment (no seeds added or sowing a diverse mix of native grassland species) in 64 780 m2 plots. The experiment will be located in naturally-assembled grasslands adjacent to BioCON experiment, and include some of the same species, allowing us to build on knowledge from these experimental monocultures and mixtures, and test whether their results and predictions can be extended to naturally-assembled grasslands and larger spatial scales.
Theme 5. NEON Biodiversity and Ecosystem Function capability. Hyperspectral imagery and derived trait maps as well as flux tower data from the NSF NEON network (Townsend NSF 1638720) will be leveraged, including hyperspectral algorithms and distribution maps of 26 traits plus uncertainties for all NEON sites (
Energy Exascale Earth System Model (E3SM) and Global Trait Diversity Maps. The land model of E3SM has been run at UMN. E3SM features subroutines to estimate the influence of nutrients and biogeochemistry on the carbon cycle. Global maps at 0.5º resolution for specific leaf area, leaf nitrogen and leaf phosphorus are available (
All Themes:
Spectral Data Repositories. The NASA-supported EcoSIS.org database for spectral measurements (not images) and ancillary measurements is maintained and curated at UW, with DOIs supplied. The UZH Specchio database includes all data made available within the agreement of the UZH open access policy.
Hyperspectral Image Processing Pipeline. Two open-source data repositories for processing hyperspectral imagery HyTools (citation) provides workflow and code to generate functional trait maps and uncertainties from atmospherically corrected hyperspectral data sources (e.g., NEON, AVIRIS, HySpex), including BRDF and topographic correction. The open-source HyPro workflow enables processing for UAS and airplane HySpex imagery, which performs orthocorrection, atmospheric correction, and smile/keystone correction to instrument radiance data. Models and related code are stored on the NASA-supported repository EcoSML.org, with DOIs. We also have access to the Laboratory Goniometer System (LAGOS) and calibration facilities of the Remote Sensing Laboratories, UZH.
BROADER IMPACTS
Education and Training
Our Institute will equip the next generation of integrative biologists with the interdisciplinary skills, capacity, perspectives, and passion required for integrative research that enhances their traditional “disciplinary” training. The Institute will provide numerous integrative training opportunities to postdoctoral associates, graduate students and undergraduates at UMN, UW, and U Maine. Postdocs will receive training at UMN, UW and U Maine; graduate students will receive training at UMN and UW; and undergraduates will receive training at UMN, UW, and U Maine. Each institution will also involve undergraduates through internal programs, as well as REUs or other NSF programs to broaden participation of diverse undergraduates. The Institute’s unifying theme and overall glue will be dual foci in spectral biology and process-based modeling, with specializations in fields including micro- and macro-evolution, plant physiological ecology, genetics, plant biochemistry and metabolomics, community ecology, ecosystem ecology, phytopathology, plant-insect and plant-microbe interactions, and global ecology. We will use a variety of training opportunities and mentoring activities, which include the following:
Training Initiatives
I. Inclusive training of diverse graduate students and postdocs. The Institute will invest significantly in training of graduate students and postdocs. The UMN, UW, and Maine teams have made extensive efforts within their graduate recruiting programs to attract underrepresented students, and the Directors and Project Leaders have strong track records of recruiting diverse graduate students and postdocs including from Hispanics, Asian Americans, international, first-generation college, and LGTBQ communities. In addition, we have assembled a diverse team of Project Leaders, including women, early career researchers, and underrepresented minorities.
II. Undergraduate research opportunities. Undergraduates will play major roles in Institute research. All investigators have strong track records training undergraduates, including recruiting and training Native American students and first generation college students, as demonstrated through prior training in the Dimensions of Biodiversity Broadening Participation program, the LTER-NASA NICE-T partnership, and the UMN and UW Undergraduate Research Experience for Undergraduates (UROP) programs.
III. Wisconsin Tribal Conservation Advisory Council (WTCAC). Our Institute is committed to fostering an inclusive research environment and training the next generation of integrative biologists to become stewards of our natural resources. Climate and land use-change pose significant threats to forest systems via stress-induced susceptibility to native and invasive pests and pathogens. The eleven Native American Tribes in Wisconsin maintain independent forestry agencies that vary in their sustainable management approaches despite similar climate-related challenges. Therefore, Tribal forests provide a unique opportunity to determine best practices to address ecological issues while contributing to the overarching aims of our Institute. The techniques and data collected from Tribal land will, in turn, provide much-needed information for Tribal forest health specialists suffering from resource and personnel limitations. To foster collaboration between UW and Tribal forestry agents, Project Leader Trowbridge has been developing an integrative training initiative with Lacey Hill-Kastern, Tribal Pest Survey Specialist with WTCAC. WTCAC is a forum for the eleven Tribes to come together and solve natural resource and conservation issues on Tribal Lands, and Hill-Kastern will foster collaboration between UW and the Tribes. Thus, together with Tribal input, we aim to develop a large project around an issue that is of interest to many—if not all—of the Tribes (e.g. oak wilt, beech leaf disease) and that involves a clade of interest to our Institute. We anticipate that the project would be overseen by WTCAC, pending approval by the Tribes and the WTCAC Board. Graduate and undergraduate students affiliated with the Institute will train with Tribal forest health specialists and learn sustainable management practices currently used to address ecological challenges. In turn, Tribal interns and students recruited through existing WTCAC initiatives (e.g. seasonal workers, pest survey program) will learn cross-disciplinary measurement techniques and data processing with affiliates of our Institute. Through this training, we also hope to recruit Native American interns into UW M.S. programs. An M.S. degree in a forestry or entomology program is necessary to obtain a Tribal forestry agency position, empowering Native American students to make critical decisions with far-reaching effects on their own lands. Bringing students, researchers, and agencies together to address shared environmental concerns will not only enhance severely understaffed Tribal conservation efforts, but will also break down barriers to information sharing between universities and the Tribes, thus enriching both the science and the students’ experiences in immeasurable ways.
IV. Annual summer workshop courses on concepts and methods. 3-4 day summer workshops will focus on specific cross-cutting themes of biological integration. In them, scientists trained in one subdiscipline will learn about concepts and methods of others, and all participants will learn how to think, observe, experiment, and model across those fields. Workshops provide a combination of skill development and training including techniques, methods in modeling and bioinformatics, and conceptual background within and across project themes. Project Leaders will be responsible for modules within the workshop, including both conceptual and technical components. Workshops will rotate annually between UMN and UW, hosted at Institute field sites or laboratories. Workshops will be open to Institute undergraduate and graduate students, postdocs, and faculty, as well as international students and postdocs from our foreign collaborators. Additional junior scientists from outside our Institute will be invited to participate to expand the impact of our training and potentially build the Institute research team.
V. Annual winter symposium on research progress. A ca. 1-day symposium will occur annually, rotating between UMN and UW, for investigators to share plans, progress and findings from their Institute work. PIs have organized similar symposia for a number of prior projects, e.g., at National Center for Ecological Analysis and Synthesis (Cavender-Bares, Isbell), Cedar Creek LTER (Hobbie, Isbell), the German Centre for Integrative Biodiversity Research (Isbell), the National Institute for Mathematical Biology and Synthesis (Cavender-Bares, Townsend, Meireles); and many others.
VI. PhysFest 2021. UMN will host PhysFest at the CCESR in 2021 (Cavender-Bares, Reich and Montgomery in collaboration with J. Nippert, C. Still and B. Helliker). The training workshop, primarily for graduate students, has been funded by NSF-IOS and is in its fourth instantiation. Building on Institute research, we will add a spectral biology component to train students in leaf level and canopy spectroscopy and interpretation, and conceptual background and application.
VII. International and domestic exchange programs. The Institute will enhance training opportunities for junior and early career scientists, harnessing national and international in-person and virtual training and collaboration opportunities at the University of Zurich in the University Research Priority Program on Global Change and Biodiversity, through the Canadian Airborne Biodiversity Observatory (CABO), and with scientists and engineers at the Jet Propulsion Laboratory. Training at the University of Zurich will focus on metabolomics, spectroscopy, remote sensing, and plant physiological modeling across scales. Training at CABO will focus on bioinformatics, theoretical community ecology, and spectroscopic methods.
VIII. Synthesis Working Groups. Two Institute synthesis working groups (undergraduates, graduate students, postdocs and other senior personnel) will use two 3-4 day workshops to develop and implement synthesis plans to integrate Institute research and work (and researchers) from outside the Institute.
IX. Special Seminar Courses. The Institute will conduct cross-institution seminars using videotechnology for both Institute and non-Institute students. These will be led by Institute investigators for both early stage (freshman) and advanced stage (upper-level undergraduate and graduate) students and address issues in biodiversity spanning genes, the tree of life, communities, ecosystems and the global biosphere.
X. Classroom-based Undergraduate Research Experiences (CURE). At both UMN and UW-Madison we will develop course-based undergraduate research experience (CURE) labs for non-major and major biology courses that provide students the opportunity to work with biodiversity data and participate in ongoing research projects. Similar existing courses have been demonstrated to increase participation of underrepresented groups in research at early career stages (
XI. Student Recruitment. Graduate students will be recruited through organizations such as Advancing Chicanos/Hispanics and Native Americans in Science (SACNAS) and by taking advantage of institution-based fellowships for underrepresented students. Undergraduate students will be recruited from faculty members’ courses, university mailing and job lists, and programs such as the North Star STEM Alliance at UMN and Undergraduate Research Scholars program at UW, programs fostering diverse scholarly communities that include students from historically underrepresented groups.
XII. Graduate student mentoring. Graduate students will be well positioned to enter the workforce for biodiversity science with a broad and integrated view afforded by the interdisciplinary training available through the proposed research. Project Leaders will mentor graduate advisees during weekly meetings to develop research questions and thesis goals, provide career guidance, discuss ideas and review progress. Lab meetings will be used to discuss primary literature and give informal presentations. Students will be encouraged to write proposals for internal and external solicitations. Extensive feedback will be provided on proposal development, manuscript preparation, and presentation skills, with opportunities to attend scientific conferences. Students will interact with diverse research groups across their own and other institutions by attending and presenting at the annual winter symposia, and attending summer workshop courses (described above). Also, students will have opportunities to engage in field work, common garden, and greenhouse experiment design and implementation, molecular bench work, plant-insect and plant-fungal work, RNAseq, bioinformatics, community ecology, physiological ecology, ecosystem ecology, and modeling within and across disciplines. Funds are dedicated to cross-institution exchange for students to learn new skills and broaden their intellectual base, encouraging a systems/integrative perspective towards understanding biodiversity. Also, graduate students will have the opportunity to mentor undergraduate students, gain teaching experience through teaching assistantships, and participate in outreach (see Outreach). Annual assessments will be performed to develop learning objectives and review progress.
XIII. Undergraduate mentoring. We have designed our undergraduate training program based on Project Leaders’ experience and peer-reviewed best practices (
XIV. Evaluation and Assessment. The efficacy of these educational and training efforts will be assessed on two main fronts. Lead by Dr. Willis in cooperation with other faculty in the Dept. of Biology Teaching and Learning (BTL) at UMN, we will assess the success of these programs (across institutions) based on pre-defined learning outcomes and learning success indicators. These efforts will focus in particular on the evaluation of the undergraduate and graduate learning programs (mentorships, seminar groups, CUREs). Second, we will assess how these programs promote inclusivity and confidence in undergraduate and graduate students, particularly, students in underrepresented minority groups. Additionally, for graduate students, we will assess how participation in these programs contributes to their career advancement and future participation in interdisciplinary research networks. Both undergraduate and graduate assessments will be performed using several pre- and post-surveys administered to students over the course of their participation in a program. Administration and analysis of both undergraduate and graduate assessments will be led by Willis in cooperation with other BTL faculty and staff.
Outreach
We will partner with the Wisconsin Tribal Conservation Association (via initiatives described above, Activity III), the Bell Museum, Minnesota’s state natural history museum (Activity XV), the Cedar Creek Ecosystem Science Reserve (CCESR) (Activity XVI), Neptune Studio’s MinuteEarth video channel (Activity XVII) and MarketScience.org (Activity XVIII) to interpret and connect the process and outcomes of our research, and the researchers themselves, with public audiences.
Diversity and inclusion. The Institute will diversify and broaden participation among team leadership, researchers, students, and trainees through a team leadership that is balanced among genders and career stages and is inclusive of researchers from underrepresented groups. Our team has a strong track record of mentoring junior scientists of diverse and underrepresented backgrounds and is committed to prioritizing diversity and inclusiveness in research and training of undergraduates, graduate students and postdocs.
Wisconsin Tribal Conservation Advisory Council. The activities associated with WTCAC are detailed above under Education and Training (III), but as described above, this effort also includes a considerable outreach component, including presentations and participation in Institute activities and workshops.
Bell Museum. Institute researchers will leverage previous collaborations with the Bell Museum on an exhibit space called “Imagine the Future.” The 1,600 square-foot exhibit area focuses on three important lessons from nature, critical for addressing the grandest challenges facing people and the planet: Thriving with Diversity, Living Within Limits, and Adapting to Change. The Thriving with Diversity section, which includes interpretive panels, casework with specimens and tools, and an interactive video kiosk, currently highlights ground-breaking biodiversity research from CCESR, and is set against a large-scale mural mosaic of spectral images from the BioDIV experiment. In Year 2 of the proposed project, Project Leaders will collaborate with the Bell Museum exhibits team to refresh Thriving with Diversity content that highlights the project’s integrative approaches to studying biodiversity across scales, with a special focus on process and outcomes. Institute researchers will assist with content development and be featured in panel text and associated interactive videos. As they are produced (see below), the three Minute Earth videos will be incorporated into the interactive video kiosk as part of the exhibit. The Bell Museum will assess visitation and experience in the updated gallery as part of the visitor experience exit survey it employs (administered via participation in COVES, Collaboration for Ongoing Visitor Experience Studies). Survey results will be assessed with Institute researchers to inform future modifications.
The Bell Museum hosts a regular public science event series called Spotlight Science, which showcases researchers working in specific areas. These have recently included chronic wasting disease, neuroscience, and plant biology. Researchers across career stages engage visitors through brief presentations and hands-on activities where the primary goal is to create conversation and opportunities for mutual learning between scientists and the public. During each year of the Institute, the Bell Museum will work with the project team to host an Institute-themed Spotlight Science. Prior to each year’s event, Amber Kastner, the Bell Museum’s public science events manager, will host a workshop for Institute researchers (with a special emphasis on graduate students and early career researchers) on best practices in public engagement and science communication. She will help researchers develop meaningful and engaging hands-on activities to be used during Spotlight Science and subsequently repurposed for other types of public engagement, including CCESR’s monthly Lunch with a Scientist series and annual Open House events. We will assess outcomes on both visitors and participating scientists (using surveys and methodologies developed by the NSF AISL-funded EvalFest team), and use the results to inform and modify programs as necessary.
Last year the Bell Museum served over 230,000 visitors from every Minnesota county and nearly every US state. This included nearly 25,000 K-12 students, over half of whom were members of underrepresented minority groups and nearly 75% were economically disadvantaged. The Bell Museum Public Engagement and Science Learning team includes informal science educators, science communicators, and exhibit and program designers who are skilled in interpreting contemporary research for, and determining its impact on, public audiences. They have specific expertise in working with researchers across career levels (from graduate students to senior scientists) and disciplines to develop exhibits, programs, and activities that engage visitors in meaningful ways.
CCESR K-12 activities for diverse learners and teacher development. CCESR is a biological field station with a legacy of ground-breaking science dating to the 1940s. CCESR hosts more than 14,000 visitors annually for hands-on science experiences. Half of these visitors are K-12 students and their teachers, the majority of whom are from public school districts with significant underrepresented minority populations. In addition, CCESR has number of programs that specifically target underserved students, including a partnership with the American Indian Education program in a local school district, a high school serving recent immigrants, a number of special education programs for young adults, and several charter schools that explicitly serve Muslim students, Hispanic/Latinx students, English Language Learners, and other underserved populations. CCESR’s education staff work closely with site scientists to develop programs that bring science to life and build a pathway of literacy and learning for all ages.
We will host a local K-12 teacher at CCESR each summer during the project. The teacher will receive a stipend to work alongside project investigators to assist with fieldwork and data collection. At the end of the summer and into the fall, CCESR education staff will assist the teacher in developing a lesson or activity related to their experience on the project. This lesson will be delivered by the teacher in their classroom and be made available to other teachers via CCESR’s website. Additionally, funding from the Institute will support the affiliated teacher to bring up to four classes of students on a field trip to CCESR to build on their in-classroom lesson. We will prioritize recruiting teachers from underrepresented groups serving in high-need school districts who have previous connections with CCESR. A new teacher partner will be selected for each summer. Dr. Caitlin Barale Potter, education and community engagement coordinator at CCESR, will oversee the teachers and coordinate the fieldwork and class visits.
MinuteEarth. To further the reach of our public engagement outside of the Upper Midwest, MinuteEarth, in cooperation with team researchers, will produce three videos about linked themes from our collective research at UMN, UW-Madison and beyond. Neptune Studio’s MinuteEarth is a popular (>300 million views) video science channel on YouTube and other platforms that has produced >190 short videos (2-3 minutes) released in English (but many also in Spanish and Portuguese) versions on YouTube, including many on biology, ecology, and global environmental change, geared to the general public audience. Co-Director Reich is a member of the MinuteEarth production team. The MinuteEarth team is expert in translating complex scientific concepts into language and visuals understandable by non-scientists, yet doing so with great scientific rigor and with an entertaining style that attracts sizeable audiences. Videos on topics such as “the economic value of biodiversity”, “the use of AI in ecological research”, “honeybee chemical defenses”, “orchid evolution and ecology”, and “the tree of life” have each been seen by hundreds of thousands, and often millions, of viewers. One video will focus on plants, light and spectra, introducing viewers to the concept of hyperspectral data in close coordination with Co-Director Townsend. The specific topics for the three videos will be developed jointly by Institute and MinuteEarth teams to optimize communication of important science, and to best represent our Institute themes, while also attracting as large an audience as possible.
Market Science. Our Science Team will further engage the public through the Market Science program at UMN, which has been running since 2014. Market Science is a collective of scientists from the University of Minnesota, and around the Twin Cities, sharing science through hands-on learning activities for kids, answering scientific questions for market goers, and creating conversations between researchers and their communities. The program involves scientists of all career stages—from students on up—who aim to connect people with science through public talks and hands-on experiences at venues including farmer’s markets and county fairs in Minnesota. We will prepare short, interactive teaching modules to present via Market Science. These modules will explain how human activities are impacting biodiversity and how we can monitor and understand these impacts through NEON and from space.
Data Management Plan
This data management plan follows best practices for data management throughout all stages of the data life-cycle according to practices outlined by DataOne, Data Carpentry and Software Carpentry.
1. Data types to be produced: Spectra from individual leaves/canopies will be stored in CSV or spreadsheet format on locally-managed and backed up to servers at their respective universities before being transferred to EcoSIS (see below), Airborne imaging spectroscopic data collected in support of this project by UW-Madison (airplane or drone) will be in ENVI format and stored on the UW Campus Computing Initiative (CCI) Cluster and distributed initially via secure website; the data collected by UZH (airplane) are stored via the Specchio server (backed up nightly). LiDAR point cloud data (LAS format) will be collected by UAV or terrestrial laser scanner, with the data transferred to locally-managed and backed-up servers at project teams’ local universities. Plant growth data and other field or lab plant measurements (e.g., chemistry, physiology, community data) will be collected at numerous field sites by investigators from all participating institutions, and will be stored as CSV or spreadsheet files in a common format, and backed up to a central server daily. High-throughput sequencing data will be collected and analyzed at each site and shared via the Open Semantic Framework (OSF) project instance. High-throughput sequencing data generated by the UMN Genomics Center is stored at the Minnesota Supercomputing Institute on Tier 2 research data for access by all project personnel. Sequencing data generated at the UZH will be maintained as described below under Storage and Backup. Metabolomics data will be stored and backed up locally on the UW-Madison CCI before dissemination. Computer code and software created during the project will be hosted publicly on GitHub, GitLab or Bitbucket. Tutorials and user documentation written for any of the tools developed for this project will be hosted and maintained on the open repositories. Tissue samples and vouchers used in this project will be obtained from existing arboreta collections. If voucher specimens are not already available, voucher specimens will be collected, prepared, and deposited at the UMN Herbarium. Tissues will be destroyed for transcriptomic and metabolomics analysis, with remaining samples stored in -80°C freezers in each PI’s Lab. Curriculum materials, including presentation slides, tutorials, code, and test data sets, will be developed for training workshops and seminars. Lessons, notes, and teaching aids will be created for K-12 classrooms. Short videos and panel text for interpretive displays will be developed for museums.
Storage and Backup: Data stored on UMN servers is backed up with snapshots taken 3x daily. 90% of storage is replicated across multiple locations (total 90 snapshots). Data at UZH is stored on the URPP-GCB Dropbox Business account which provides version control, as well as file recovery for 180 days, and additionally backed up to a local server daily; files on the local server are backed up daily to a separate hard disk and additionally archived to two separate magnetic tapes. All UW-Madison data are stored on the Campus Computing Initiative (CCI) servers and backed up to tape nightly. In addition, the Townsend lab has a server on which working data are incrementally backed up hourly.
2. Data standards to be used: Data generated by the project will be retained in script-readable formats: CSV or spreadsheet file for numerical data; GeoTIFF, HDF5 or ENVI for imagery; and NetCDF, shapefiles and KMLs for GIS data. Upon initiation of the CCESR, a standard format will be distributed for metadata and metadata entries will be published in a searchable interface on the Institute website upon publication of the associated datasets, including a link to the published dataset, to facilitate data re-use. To assure quality control and reproducibility of the project outputs, two secure cloud-based platforms will allow the distributed project team to organize, annotate, and document the data. Open Science Framework (osf.io) will be used by the project team to collaborate and create shared documentation via the wiki function and the connected shared drives. OSF will ensure good communication between the team members and act as a collaborative electronic lab notebook for disparate data types and sites. Fulcrum (www.fulcrumapp.com) will be used by the project team to record site information (including metadata) for all leaf spectral data, enabling interoperability among all participants through consistent cataloguing and descriptions for all participants. Fulcrum data are instantaneously backed up to the cloud when connected, and will be exported for backup and extraction when the project is complete.
3. Data management roles and responsibilities: Information managers at UMN and UW-Madison will coordinate data management activities in consultation with the Director, Co-Directors, Managing Directors and the science team. All science team members will be required to use Fulcrum for lab and field data collection that will be synced on a common server. Project Leaders will develop data entry forms and approve data submissions. If any member of the Institute directors, project leaders, or information managers leaves the project, their data management responsibilities will be transferred to one of the remaining directors, either until the end of the project or a replacement is named.
4. Data dissemination methods and access: Leaf and plot hyperspectral data and any associated measurements/descriptions (e.g., species information, chemical/physiological measurements, metabolomics etc.) will be publicly searchable and available through EcoSIS.org, a NASA-supported repository hosted on Amazon Web Services. EcoSIS supplies DOIs for all contributions and is managed and curated by co-PI Townsend at UW-Madison. EcoSIS is backed up daily. Airborne imaging spectroscopic and LiDAR data collected by our project team will be publicly searchable and downloadable from NASA Distributed Active Archive Centers (DAACs). Image data collected by third parties (e.g., NASA, NEON) will be available through their portals. Plant growth data from experiments at Cedar Creek Ecosystem Science Reserve (CCESR) will be made publicly available via the Environmental Data Initiative (EDI) with links to the CDR LTER website, an NSF-funded resource for long-term public access and preservation. Data collected in Minnesota but not at CCESR will be stored on shared, locally managed servers at UMN and deposited into the UMN Libraries’ Data Repository for the University of Minnesota (DRUM), an open access core-trust seal certified institutional data repository (more information on DRUM at 6. Data archiving below). High-throughput sequencing data of individual plants will be shared and distributed via the NCBI Sequence Read Archive (SRA). Phylogenetic datasets, including trees and analysis information, will be deposited to the OpenTree project, allowing for further annotation and sharing of the results. In addition, analysis files for publications, such as sequence alignments, climatic niche and metabolite data matrices, any other source data files, and various associated statistical analyses will be permanently stored in the Dryad Digital Repository. Computer code and software will be developed under the GPL (Gnu Public License) or similar certified open-source license (as defined by the Open Source Initiative). All code for processing of hyperspectral data (leaf measurements and image data) will be distributed through EcoSML.org, a NASA supported searchable open-source code repository linked to GitHub, GitLab and BitBucket that supplies DOIs for all contributions. EcoSML is managed and curated by Co-PI Townsend at UW-Madison. All code repositories will be actively maintained and updated by the authors after publication. In addition, the version used for the publication will be provided in the supplemental material of all publications produced. Tissue samples will be freely available for nonprofit research uses upon permission from the original tissue provider after completion of the project. Curriculum materials will be deposited in DRUM for public availability with the exception of K-12 classroom materials, which will be made publicly available on the CCESR website.
5. Data policies: Data access will be restricted to the project personnel during collection and analysis and released immediately upon publication of research findings. If data are archived prior to publication, the data will be embargoed until date of publication. Data will be released under a Creative Commons BY 2.0 License requesting attribution for reuse.
6. Data archiving: The archive repositories described above ensure long-term preservation and access. All software code hosted on GitHub will be archived in the GitHub Archive Program. Software code hosted on BitBucket will be submitted to the Software Heritage archive (softwareheritage.org). Digital data not already in one of these archives at project’s end will be deposited in DRUM. All archived data will include the appropriate documentation, metadata, and code to facilitate reuse.
Funding program
US National Science Foundation (NSF) Award Number: 2021898
Division of Biological Infrastructure (DBI) Biology Integration Institutes
Grant title
BII-Implementation: The causes and consequences of plant biodiversity across scales in a rapidly changing world
Hosting institution
University of Minnesota
References
- A trait-based ecosystem model suggests that long-term responsiveness to rising atmospheric CO2 concentration is greater in slow-growing than fast-growing plants.Functional Ecology27(4):1011‑1022. https://doi.org/10.1111/1365-2435.12102
- Elevated carbon dioxide is predicted to promote coexistence among competing species in a trait-based model.Ecology and Evolution5(20):4717‑4733. https://doi.org/10.1002/ece3.1733
- Spectranomics: Emerging science and conservation opportunities at the interface of biodiversity and remote sensing.Global Ecology and Conservation8:212‑219. https://doi.org/10.1016/j.gecco.2016.09.010
- Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation.Science355(6323):385‑389. https://doi.org/10.1126/science.aaj1987
- Belowground biodiversity and ecosystem functioning.Nature515(7528):505‑511. https://doi.org/10.1038/nature13855
- An Overview of AVIRIS-NG Airborne Hyperspectral Science Campaign Over India.Current Science116(7). https://doi.org/10.18520/cs/v116/i7/1082-1088
- Variational Inference: A Review for Statisticians.Journal of the American Statistical Association112(518):859‑877. https://doi.org/10.1080/01621459.2017.1285773
- A test of the hierarchical model of litter decomposition.Nature Ecology & Evolution1(12):1836‑1845. https://doi.org/10.1038/s41559-017-0367-4
- Poaceae genomes: Going from unattainable to becoming a model clade for comparative plant genomics.Plant Physiology149(1):111‑116. https://doi.org/10.1104/pp.108.128926
- Mapping local and global variability in plant trait distributions.Proceedings of the National Academy of Sciences114(51). https://doi.org/10.1073/pnas.1708984114
- The influence of functional diversity on terrestrial carbon uptake.In Prep.
- Biodiversity loss and its impact on humanity.Nature486:59‑67. https://doi.org/10.1038/nature11148
- Associations of leaf spectra with genetic and phylogenetic variation in oaks: prospects for remote detection of biodiversity.Remote Sensing8(3). https://doi.org/10.3390/rs8030221
- Harnessing plant spectra to integrate the biodiversity sciences across biological and spatial scales.American Journal of Botany104(7):1‑4. https://doi.org/10.3732/ajb.1700061
- The role of diversification in community assembly of the oaks (Quercus L.) across the continental U.S.American Journal of Botany105(3):565‑586. https://doi.org/10.1002/ajb2.1049
- Diversification, adaptation, and community assembly of the American oaks (Quercus), a model clade for integrating ecology and evolution.New Phytologist221(2):669‑692. https://doi.org/10.1111/nph.15450
- SteadyCom: Predicting microbial abundances while ensuring community stability.PLOS Computational Biology13(5). https://doi.org/10.1371/journal.pcbi.1005539
- Ecological Niches: Linking classical and contemporary approaches.University of Chicago Press,Chicago.
- mvmorph: an r package for fitting multivariate evolutionary models to morphometric data.Methods in Ecology and Evolution6(11):1311‑1319. https://doi.org/10.1111/2041-210X.12420
- The International Land Model Benchmarking (ILAMB) System: Design, Theory, and Implementation.Journal of Advances in Modeling Earth Systems10(11):2731‑2754. https://doi.org/10.1029/2018MS001354
- Using foliar spectral properties to assess the effects of drought on plant water potential.Tree Physiology37(11):1582‑1591. https://doi.org/10.1093/treephys/tpx106
- Intraspecific genetic variation of a Fagus sylvatica population in a temperate forest derived from airborne imaging spectroscopy time series.Ecology and Evolution10(14):7419‑7430. https://doi.org/10.1002/ece3.6469
- Multiple elements of soil biodiversity drive ecosystem functions across biomes.Nature Ecology & Evolution4(2):210‑220. https://doi.org/10.1038/s41559-019-1084-y
- Predicting ecosystem stability from community composition and biodiversity.Ecology Letters16(5):617‑625. https://doi.org/10.1111/ele.12088
- Using imaging spectroscopy to detect variation in terrestrial ecosystem productivity across a water-stressed landscape.Ecological Applications28(5):1313‑1324. https://doi.org/10.1002/eap.1733
- Elevated CO2 does not increase eucalypt forest productivity on a low-phosphorus soil.Nature Climate Change7(4):279‑282. https://doi.org/10.1038/nclimate3235
- Students as ecologists: Strategies for successful mentorship of undergraduate researchers.Ecology and Evolution9(8):4316‑4326. https://doi.org/10.1002/ece3.5090
- Improving Underrepresented Minority Student Persistence in STEM.CBE—Life Sciences Education15(3):es5. https://doi.org/10.1187/cbe.16-01-0038
- Spectral differentiation of oak wilt from foliar fungal disease and drought is correlated with physiological changes.Tree Physiology40(3):377‑390. https://doi.org/10.1093/treephys/tpaa005
- An integrated hyperspectral imaging and genome-wide association analysis platform provides spectral and genetic insights into the natural variation in rice.Sci. Rep.7(1):4401. https://doi.org/10.1038/s41598-017-04668-8
- PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle.Remote Sens. Environ.193:204‑215. https://doi.org/10.1016/j.rse.2017.03.004
- WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas.Int. J. Climatol.37(12):4302‑4315. https://doi.org/10.1002/joc.5086
- Missing pieces to modeling the Arctic-Boreal puzzle.Environmental Research Letters13(2). https://doi.org/10.1088/1748-9326/aa9d9a
- ECOSTRESS: NASA’s next generation mission to measure evapotranspiration from the International Space Station.Water Resources Research56(4):e2019WR02605. https://doi.org/10.1029/2019WR026058
- Exploring the Earth System with Imaging Spectroscopy.Springer,Cham, Switzerland.
- Land mosaics: the ecology of landscapes and regions.Cambridge University Press,Cambridge, UK.
- Consideration of scale in remote sensing of biodiversity. In:Remote Sensing of Plant Biodiversity.Springer Verlag,New York.
- Shifting relative importance of climatic constraints on land surface phenology.Environmental Research Letters13(2):024025. https://doi.org/10.1088/1748-9326/aaa17b
- Data Analysis using Regression and Multilevel/Hierarchical Models.Cambridge University Press,New York.
- Detecting prairie biodiversity with airborne remote sensing.Remote Sensing of Environment221:38‑49. https://doi.org/10.1016/j.rse.2018.10.037
- Multi-task Sparse Structure Learning with Gaussian Copula Models.Journal of Machine Learning Research17(33):1‑30. URL: http://jmlr.org/papers/v17/15-215.html
- Scaling-up biodiversity-ecosystem functioning research.Ecology Letters23(4):757‑776. https://doi.org/10.1111/ele.13456
- Species richness and traits predict overyielding in stem growth in an early-successional tree diversity experiment.Ecology98(10):2601‑2614. https://doi.org/10.1002/ecy.1958
- Macrosystems ecology: understanding ecological patterns and processes at continental scales.Frontiers in Ecology and the Environment12(1):5‑14. https://doi.org/10.1890/130017
- The "wake-sleep" algorithm for unsupervised neural networks.Science268(5214):1158‑1161. https://doi.org/10.1126/science.7761831
- Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity.New Phytologist217(1):439‑452. https://doi.org/10.1111/nph.14773
- Genomic landscape of the global oak phylogeny.New Phytologist226(4):1198‑1212. https://doi.org/10.1111/nph.16162
- A comparison of Dynamic Habitat Indices derived from different MODIS products as predictors of avian species richness.Remote Sensing of Environment195:142‑152. https://doi.org/10.1016/j.rse.2017.04.018
- Simple rules for interspecific dominance in systems with exploitative and apparent competition.The American Naturalist144(5):741‑771. https://doi.org/10.1086/285705
- Review of optical-based remote sensing for plant trait mapping.Ecological Complexity15:1‑16. https://doi.org/10.1016/j.ecocom.2013.06.003
- Effects of Biodiversity on Ecosystem Functioning: A Consensus of Current Knowledge.Ecological Monographs75(1):3‑35. https://doi.org/10.1890/04-0922
- Implications of improved representations of plant respiration in a changing climate.Nature Communications8(1):1602. https://doi.org/10.1038/s41467-017-01774-z
- Summary for policymakers of the global assessment report on biodiversity and ecosystem services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services.IPBES secretariat, Bonn, Germany.
- Linking the influence and dependence of people on biodiversity across scales.Nature546(7656):65‑72. https://doi.org/10.1038/nature22899
- PROSPECT plus SAIL models: A review of use for vegetation characterization.Remote Sensing of Environment113(S1):S56‑S66. https://doi.org/10.1016/j.rse.2008.01.026
- Leaf Optical Properties.Cambridge University Press,New York.
- Monitoring plant functional diversity from space.Nature Plants2(3):16024. https://doi.org/10.1038/nplants.2016.24
- The fate of carbon in a mature forest under carbon dioxide enrichment.Nature580(7802):227‑231. https://doi.org/10.1038/s41586-020-2128-9
- TRY plant trait database - enhanced coverage and open access.Glob. Chang. Biol.26(1):119‑188. https://doi.org/10.1111/gcb.14904
- An Introduction to Variational Autoencoders.Foundations and Trends® in Machine Learning12(4):307‑392. https://doi.org/10.1561/2200000056
- Solanaceae — a model for linking genomics with biodiversity.Comparative and Functional Genomics5(3):285‑291. https://doi.org/10.1002/cfg.393
- Quantile Regression.Cambridge CoreURL: /core/books/quantile-regression/C18AE7BCF3EC43C16937390D44A328B1
- Community-wide consequences of variation in photoprotective physiology among prairie plants.Photosynthetica56:455‑467. https://doi.org/10.1007/s11099-018-0777-9
- Partitioning plant spectral diversity into alpha and beta components.Ecology Letters23(2):370‑380. https://doi.org/10.1111/ele.13429
- Linking Earth Observation and taxonomic, structural and functional biodiversity: Local to ecosystem perspectives.Ecological Indicators70:317‑339. https://doi.org/10.1016/j.ecolind.2016.06.022
- Linking Remote Sensing and Geodiversity and Their Traits Relevant to Biodiversity—Part I: Soil Characteristics.Remote Sensing11(20):2356. https://doi.org/10.3390/rs11202356
- Deep learning.Nature521(7553):436‑444. https://doi.org/10.1038/nature14539
- Community assembly and the functioning of ecosystems: how metacommunity processes alter ecosystems attributes.Ecology98(4):909‑919. https://doi.org/10.1002/ecy.1697
- The metacommunity concept: a framework for multi-scale community ecology.Ecology Letters7(7):601‑613. https://doi.org/10.1111/j.1461-0248.2004.00608.x
- Positive biodiversity-productivity relationship predominant in global forests.Science354(6309). https://doi.org/10.1126/science.aaf8957
- Memote: A community driven effort towards a standardized genome-scale metabolic model test suite.bioRxivhttps://doi.org/10.1101/350991
- Origin of angiosperms and the puzzle of the Jurassic gap.Nat Plants5(5):461‑470. https://doi.org/10.1038/s41477-019-0421-0
- A Global, 0.05-Degree Product of Solar-Induced Chlorophyll Fluorescence Derived from OCO-2, MODIS, and Reanalysis Data.Remote Sensing11(5):517. https://doi.org/10.3390/rs11050517
- Partitioning selection and complementarity in biodiversity experiments.Nature412:72‑76. https://doi.org/10.1038/35083573
- Biodiversity as spatial insurance in heterogeneous landscapes.Proceedings of the National Academy of Sciences100(22):12765‑12770. https://doi.org/10.1073/pnas.2235465100
- Future global productivity will be affected by plant trait response to climate.Scientific Reports8(1):2870. https://doi.org/10.1038/s41598-018-21172-9
- Imaging spectroscopy links aspen genotype with below-ground processes at landscape scales.Philosophical Transactions of the Royal Society of London B: Biological Sciences369(1643):20130194. https://doi.org/10.1098/rstb.2013.0194
- Variability and Uncertainty Challenges in Scaling Imaging Spectroscopy Retrievals and Validations from Leaves Up to Vegetation Canopies.Surveys in Geophysics40(3):631‑656. https://doi.org/10.1007/s10712-019-09534-y
- Lessons learned from Spectranomics: wet tropical forests. In:Remote Sensing of Plant Biodiversity.Springer,New York.
- Linking leaf spectra to the plant tree of life. In:Remote Sensing of Plant Biodiversity.Springer,17pp. https://doi.org/10.1007/978-3-030-33157-3_7
- Leaf reflectance spectra capture the evolutionary history of seed plants.New Phytologist228(2):485‑493. https://doi.org/10.1111/nph.16771
- Detecting drought stress in soybean plants using hyperspectral fluorescence imaging.Journal of Biosystems Engineering40(4):335‑344. URL: https://www.e-sciencecentral.org/articles/SC000013860
- A Novel Approach to Assess Salt Stress Tolerance in Wheat Using Hyperspectral Imaging.Front. Plant Sci.9(1182). https://doi.org/10.3389/fpls.2018.01182
- A methodology to derive global maps of leaf traits using remote sensing and climate data.Remote Sensing of Environment218:69‑88. https://doi.org/10.1016/j.rse.2018.09.006
- Biodiversity promotes primary productivity and growing season lengthening at the landscape scale.Proceedings of the National Academy of Sciences114(38):10160‑10165. https://doi.org/10.1073/pnas.1703928114
- Terrestrial land-cover type richness is positively linked to landscape-level functioning.Nature Communications11(1). https://doi.org/10.1038/s41467-019-14002-7
- Canopy nitrogen, carbon assimilation, and albedo in temperate and boreal forests: Functional relations and potential climate feedbacks.Proceedings of the National Academy of Sciences105(49):19335‑19340. https://doi.org/10.1073/pnas.0810021105
- What is flux balance analysis?Nature Biotechnology28(3):245‑248. https://doi.org/10.1038/nbt.1614
- Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being.Science355(6332). https://doi.org/10.1126/science.aai9214
- Fire frequency drives decadal changes in soil carbon and nitrogen and ecosystem productivity.Nature553(7687):194‑198. https://doi.org/10.1038/nature24668
- Using remote sensing for modeling and monitoring species distributions. In:Remote Sensing of Plant Biodiversity.Springer,New York. [ISBN9783030331566].
- Emergent properties from organisms to ecosystems: towards a realistic approach.Biological Reviews80(3):403‑411. https://doi.org/10.1017/S146479310500672X
- The Dynamic Habitat Indices (DHIs) from MODIS and global biodiversity.Remote Sensing of Environment222:204‑214. https://doi.org/10.1016/j.rse.2018.12.009
- Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models.Remote Sensing of Environment239:111626. https://doi.org/10.1016/j.rse.2019.111626
- Gaussian Processes for Machine Learning.The MIT Press,Cambridge, Mass. URL: https://www.amazon.com/Gaussian-Processes-Learning-Adaptive-Computation/dp/026218253X [ISBN978-0-262-18253-9]
- Key canopy traits drive forest productivity.Proceedings of the Royal Society B: Biological Sciences279(1736):2128‑2134. https://doi.org/10.1098/rspb.2011.2270
- Decade-long soil nitrogen constraint on the CO2 fertilization of plant biomass.Nature Climate Change3(3):278‑282. https://doi.org/10.1038/nclimate1694
- The world-wide ‘fast–slow’ plant economics spectrum: a traits manifesto.Journal of Ecology102(2):275‑301. https://doi.org/10.1111/1365-2745.12211
- Biogeographic variation in evergreen conifer needle longevity and impacts on boreal forest carbon cycle projections.Proceedings of the National Academy of Sciences111(38):13703‑13708. https://doi.org/10.1073/pnas.1216054110
- Geographic range predicts photosynthetic and growth response to warming in co-occurring tree species.Nature Climate Change5(2):148‑152. https://doi.org/10.1038/nclimate2497
- Boreal and temperate trees show strong acclimation of respiration to warming.Nature531(7596):633‑636. https://doi.org/10.1038/nature17142
- Unexpected reversal of C3 versus C4 grass response to elevated CO2 during a 20-year field experiment.Science360(6386):317‑320. https://doi.org/10.1126/science.aas9313
- Effects of climate warming on photosynthesis in boreal tree species depend on soil moisture.Nature562(7726):263‑267. https://doi.org/10.1038/s41586-018-0582-4
- Linking plant and ecosystem functional biogeography.Proceedings of the National Academy of Sciences111(38). https://doi.org/10.1073/pnas.1216065111
- Design and performance of combined infrared canopy and belowground warming in the B4WarmED (Boreal Forest Warming at an Ecotone in Danger) experiment.Global Change Biology21(6):2334‑2348. https://doi.org/10.1111/gcb.12855
- From local to regional: Functional diversity in differently managed alpine grasslands.Remote Sensing of Environment236:111415. https://doi.org/10.1016/j.rse.2019.111415
- Microbial carbon use efficiency predicted from genome-scale metabolic models.Nature Communications10(1):3568. https://doi.org/10.1038/s41467-019-11488-z
- Fossils matter: improved estimates of divergence times in Pinus reveal older diversification.BMC Evol. Biol.17(1):95. https://doi.org/10.1186/s12862-017-0941-z
- Flux towers in the sky: global ecology from space.New Phytologist224(2):570‑584. https://doi.org/10.1111/nph.15934
- Prospects and pitfalls for spectroscopic remote sensing of biodiversity at the global scale. In:Remote Sensing of Plant Biodiversity.Springer,New York.
- Mapping functional diversity from remotely sensed morphological and physiological forest traits.Nature Communications8(1):1441. https://doi.org/10.1038/s41467-017-01530-3
- Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function.Nature Ecology & Evolution2(6):976‑982. https://doi.org/10.1038/s41559-018-0551-1
- Recent breakthroughs in metabolomics promise to reveal the cryptic chemical traits that mediate plant community composition, character evolution and lineage diversification.The New Phytologist214(3):952‑958. https://doi.org/10.1111/nph.14438
- A protocol for high-throughput, untargeted forest community metabolomics using mass spectrometry molecular networks.Applications in Plant Sciences6(3):1‑13. https://doi.org/10.1002/aps3.1033
- Remotely estimating photosynthetic capacity, and its response to temperature, in vegetation canopies using imaging spectroscopy.Remote Sensing of Environment167:78‑87. https://doi.org/10.1016/j.rse.2015.05.024
- Scaling functional traits from leaves to canopies. In:Remote Sensing of Plant Biodiversity.Springer,New York.
- Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties.Ecological Applications25(8):2180‑2197. https://doi.org/10.1890/14-2098.1
- Constructing a broadly inclusive seed plant phylogeny.Am. J. Bot.105(3):302‑314. https://doi.org/10.1002/ajb2.1019
- ISS observations offer insights into plant function.Nature Ecology & Evolution1(7):0194. https://doi.org/10.1038/s41559-017-0194
- Surprising lack of sensitivity of biochemical limitation of photosynthesis of nine tree species to open-air experimental warming and reduced rainfall in a southern boreal forest.Global Change Biology26(2):746‑759. https://doi.org/10.1111/gcb.14805
- Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance.Sci. Total Environ.578:90‑99. https://doi.org/10.1016/j.scitotenv.2016.08.014
- Reduced feeding activity of soil detritivores under warmer and drier conditions.Nature Climate Change8(1):75‑78. https://doi.org/10.1038/s41558-017-0032-6
- Soil microbial, nematode, and enzymatic responses to elevated CO2, N fertilization, warming, and reduced precipitation.Soil Biology and Biochemistry135:184‑193. https://doi.org/10.1016/j.soilbio.2019.04.020
- Simulating functional diversity of European natural forests along climatic gradients.Journal of Biogeography47(5):1069‑1085. https://doi.org/10.1111/jbi.13809
- Resource Competition and Community Structure.Princeton University Press
- Utilizing top-down hyperspectral imaging for monitoring genotype and growth conditions in maize.BioRxivhttps://doi.org/10.1101/2020.01.21.914069
- Retrieval of foliar information about plant pigment systems from high resolution spectroscopy.Remote Sensing of Environment113:S67‑S77. https://doi.org/10.1016/j.rse.2008.10.019
- A novel Bayesian method for inferring and interpreting the dynamics of adaptive landscapes from phylogenetic comparative data.Syst. Biol.63(6):902‑918. https://doi.org/10.1093/sysbio/syu057
- Assessing durum wheat ear and leaf metabolomes in the field through hyperspectral data.The Plant Journal102(3):615‑630. https://doi.org/10.1111/tpj.14636
- Bioclimatic variables derived from remote sensing: assessment and application for species distribution modelling.Methods in Ecology and Evolution5(10):1033‑1042. https://doi.org/10.1111/2041-210X.12264
- Influence of species richness, evenness, and composition on optical diversity: A simulation study.Remote Sensing of Environment211:218‑228. https://doi.org/10.1016/j.rse.2018.04.010
- Remote sensing of terrestrial plant biodiversity.Remote Sensing of Environment231:111218. https://doi.org/10.1016/j.rse.2019.111218
- Mapping foliar functional traits and their uncertainties across three years in a grassland experiment.Remote Sensing of Environment221:405‑416. https://doi.org/10.1016/j.rse.2018.11.016
- Foliar functional traits from imaging spectroscopy across biomes in the eastern North America.New Phytologist228(2):494‑511. https://doi.org/10.1111/nph.16711
- Spatial complementarity in tree crowns explains overyielding in species mixtures.Nature Ecology & Evolution1(4):0063. https://doi.org/10.1038/s41559-016-0063
- Remote spectral detection of biodiversity effects on forest biomass.Nature Ecology & Evolution5:46‑54. https://doi.org/10.1038/s41559-020-01329-4
- Basic and extensible post-processing of eddy covariance flux data with REddyProc.Biogeosciences15(16):5015‑5030. https://doi.org/10.5194/bg-15-5015-2018
- A new generation of the United States National Land Cover Database: Requirements, research priorities, design, and implementation strategies.ISPRS Journal of Photogrammetry and Remote Sensing146:108‑123. https://doi.org/10.1016/j.isprsjprs.2018.09.006
- An efficient field and laboratory workflow for plant phylotranscriptomic projects.Appl. Plant Sci.5(3):160012. https://doi.org/10.3732/apps.1600128
- High-Throughput Phenotyping of Maize Leaf Physiological and Biochemical Traits Using Hyperspectral Reflectance.Plant Physiol.173(1):614‑626. https://doi.org/10.1104/pp.16.01447
- High-Resolution Global Contiguous SIF of OCO-2.Geophysical Research Letters46(3):1449‑1458. https://doi.org/10.1029/2018GL081109