|
Research Ideas and Outcomes : Workshop Report
|
|
Corresponding author: Katja C. Seltmann (enicospilus@gmail.com), Deborah L. Paul (dpaul@fsu.edu)
Received: 14 Dec 2018 | Published: 17 Dec 2018
© 2018 Katja Seltmann, Sara Lafia, Deborah Paul, Shelley James, David Bloom, Nelson Rios, Shari Ellis, Una Farrell, Jessica Utrup, Michael Yost, Edward Davis, Rob Emery, Gary Motz, Julien Kimmig, Vaughn Shirey, Emily Sandall, Daniel Park, Christopher Tyrrell, R. Sean Thackurdeen, Matthew Collins, Vincent O'Leary, Heather Prestridge, Christopher Evelyn, Ben Nyberg
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Seltmann K, Lafia S, Paul D, James S, Bloom D, Rios N, Ellis S, Farrell U, Utrup J, Yost M, Davis E, Emery R, Motz G, Kimmig J, Shirey V, Sandall E, Park D, Tyrrell C, Thackurdeen R, Collins M, O'Leary V, Prestridge H, Evelyn C, Nyberg B (2018) Georeferencing for Research Use (GRU): An integrated geospatial training paradigm for biocollections researchers and data providers. Research Ideas and Outcomes 4: e32449. https://doi.org/10.3897/rio.4.e32449
|
|
Abstract
Georeferencing is the process of aligning a text description of a geographic location with a spatial location based on a geographic coordinate system. Training aids are commonly created around the georeferencing process to disseminate community standards and ideas, guide accurate georeferencing, inform users about new tools, and help users evaluate existing geospatial data. The Georeferencing for Research Use (GRU) workshop was implemented as a training aid that focused on the creation and research use of geospatial coordinates, and included both data researchers and data providers, to facilitate communication between the groups. The workshop included 23 participants with a wide background of expertise ranging from students (undergraduate and graduate), professors, researchers and educators, scientific data managers, natural history collections personnel, and spatial analyst specialists. The conversations and survey results from this workshop demonstrate that it is important to provide opportunities for biocollections data providers to interact directly with the researchers using the data they produce and vice versa.
Keywords
GIS, Geospatial data, Natural history collections, Biocollections, Workshop report, Data quality, iDigBio, Georeferencing, GeoLocate, QGIS
Introduction
Scientific knowledge relating to our environment, human health, climate change and global ecosystems increasingly requires the creation and evaluation of diverse and varied datasets, warranting training to raise the data competencies of researchers and data providers (
Background: Importance of biocollections georeferenced data
Information about the distribution of biological organisms is used to investigate many current global health and human services such as clean water, land preservation and restoration, disease prevention, food safety and security, agricultural pests, drought, land use management, urban planning, and the effects of climate change (
Georeferenced locality data are widely made available through biodiversity data aggregators such as Atlas of Living Australia (ALA), the Global Biodiversity Information Facility (GBIF), Integrated Digitized Biocollections (iDigBio), and VertNet in a digital format. Digital data records for more than 115 million physical specimens have been shared (
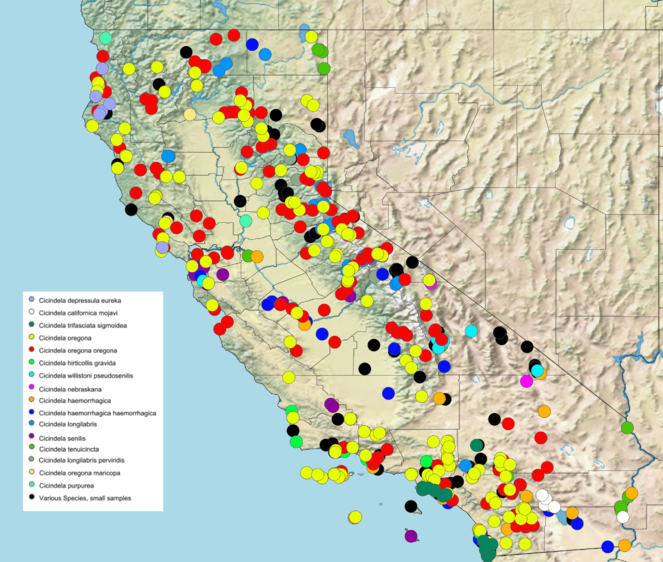
Map created using SimpleMappr (
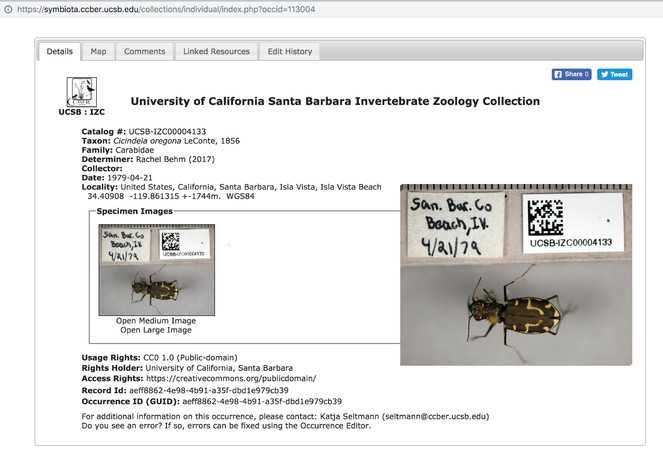
This specimen record is an example from the University of California Collection Network Symbiota Portal. The large image is an edit of the record to include a medium size version of the image for easier viewing in this article. The portal software is open source and it is freely available for reuse through the Symbiota GitHub repository. The image is an example of a specimen record that includes an image of the specimen with label data. The image is contributed by the UCSB Invertebrate Zoology Collection at the Cheadle Center for Biodiversity and Ecological Restoration. The usage rights for the image is Creative Commons 0 (public domain).
Rationale: Raising data competencies for researchers and data providers
The data captured from biocollection specimens are typically managed by collections staff who care for specimens and digitize the data associated with those specimens, ultimately making the data available for research. This creates a scenario where the people creating the datasets are often not the same people who use the data in research, which creates a need to provide venues of communication and foster understanding between these stakeholders about the implications of each other’s' methods on research products (
History: Background and provenance of recent efforts in biocollection digitization
Recognizing the value of biocollections for research, education, and society, a diverse group of scientists outlined a coordinated effort they envisioned as a Networked Integrated Biocollections Alliance (NIBA), which resulted in strategic and implementation plans for the digitization of US collections information (
As part of this initiative, the iDigBio Georeferencing Working Group (GWG) was formed to support the biocollections community to implement best practices in the improvement and maintenance of critical location data. The GWG benefits from the contributions from many, including GEOLocate, VertNet, TCNs, and earlier projects (e.g., Georeferencing.org, MaNIS, FishNet2, ORNIS and HerpNET).
In response to biocollection georeferencing needs, the GWG offered two Train-the-Tainers workshops in 2012 and 2013. These workshops were aimed at training biodiversity and collections professionals to use best practices for georeferencing (
Training provided by the iDigBio project initially focused on community-derived digitization best practice discovery and documentation but has increasingly incorporated innovative use of scientific collections data for research into workshops and symposia design. The iDigBio Georeferencing Working Group (GWG) saw the need to offer a georeferencing workshop that would combine best practices for historical and new data with new lessons on evaluating the resulting biocollections data for their fitness for research and other downstream use.
Georeferencing for Research Use (GRU) Workshop Design and Implementation
To design the Georeferencing for Research Use workshop, the GWG first reviewed materials used in prior workshops (e.g., Train-the-Trainers, Field to Database) to determine what lessons about the evaluation of geospatial data should be integrated into the course curriculum. Additional content was then included by the course instructors from Biodiversity Informatics Training Curriculum (BITC), Cheadle Center for Biodiversity and Ecological Restoration (CCBER), and University of California, Santa Barbara National Center for Ecological Analysis and Synthesis (NCEAS) and iDigBio. A specimen dataset downloaded directly from iDigBio was used as the example dataset (Suppl. material
Topic choice and time spent on specialized topics was in part guided by the applicants' expectations of the course. Using an online form that called for participation and evaluated expectations of participants (Suppl. material
Participants were asked to prepare in advance for the workshop. Pre-workshop assignments included a review of materials for a sufficient level of understanding of the fundamental principles of GIS and best practice for biocollection georeferenced data, such as projection information and coordinate reference systems.
The GRU workshop was held 4-7 October 2016 in Santa Barbara, California, hosted by the NCEAS and CCBER. The first two days of the workshop training provided a summary of biocollection georeferencing and data standards, legacy collection data issues, and best practices for the creation of new locality (geospatial) data in an effort to avoid increasing the legacy-data backlog. The final two days encompassed strategies for data standardization before research use, such as the use of spreadsheets and OpenRefine (http://openrefine.org/) software to evaluate data and to select, adjust, and remove data as appropriate, and the visualization of common geolocation issues using QGIS and other tools.
Workshop Objectives
The major objectives of the workshop were to gain feedback from the participants on research data needs for the future and to enhance and improve the quality of collections geospatial data. Objectives of the 4-day workshop included:
-
Demonstration of tools (hardware and software) and geospatial data standards, especially as relating to the Darwin Core standard.
-
Discussion of best practices for data repositories (e.g., obstacles and minimization of data loss).
-
How to evaluate already georeferenced data for quality, or the fitness of the data for a specific use (fitness-for-use).
-
Current tools for visualization and evaluation.
-
Introduction of open-source QGIS software and selected plug-ins to participants to demonstrate data visualization methods.
-
Sharing best practices for researchers for in-the-field creation of new locality data.
-
Gain insight into the challenges faced by researchers for georeferencing through shared research experiences using biocollection geospatial data.
-
Gain insight into the challenges data and collection managers experience in generating and managing georeferenced data.
-
Gain input from participants about their needs and learning experience during the workshop.
-
Gain an understanding of how participants use their learning for research and other present and future research and curation of collections and data needs.
Participant demographics
The results presented here represent views from our participants (23 persons) who were selected for their interest in georeferencing and biocollections at the time that they registered for the workshop. All but one of the participants were affiliated with U.S institutions. The participants represented a cross-section of users of biocollections data that include students (graduate and undergraduate), professors, collections managers, curators, an agriculture specialist, and data managers.
Results and Outcomes
In total, 42 applications were received in response to the open call for participants. Twenty-three participants were accepted, with priority given to persons who had not participated in prior iDigBio workshops, and whose expectations best matched the proposed workshop goals and scope. Participants primarily self-identified as researchers or research students (14 participants) who use biocollections spatial data in their research projects, or as data/collection managers (9 participants). Several participants also self-identified as having multiple career needs for participating in the training, for example, a researcher who is also managing a biocollection, or a data manager who is also a student. The combined audience fostered collaboration and understanding across these different domains.
Conversations around GIS, QGIS, and other tools
Workshop participants self-identified as a researcher, data manager, GIS expert, or some combination of identities. The needs of each of these groups for data management and georeferencing skills were similar, yet each group had particular goals. For example, researchers were interested in the functionality of OpenRefine software for standardizing biodiversity data sets, new tools for efficient field data collection, such as the Biocode Field Information Management System, and efficient georeferencing of large datasets with high-throughput analyses of specimen coordinate accuracy. There was a strong desire for efficient and accurate georeferencing of biocollections to create "research ready" data that is available for complex, data-driven analyses (
Workshop participants with a strong GIS background were primarily interested in learning more about the current practices of researchers and data managers in data cleaning, georeferencing, and spatial analysis to identify areas of opportunity for improving the ability of domain experts to assess data quality and gain new insights to visualize their data spatially. The QGIS tutorial was modified in response to the interest in new skills expressed by participants in the first two days of the workshop. For example, a desire to know how to leverage coupled spatial data layers to perform a spatial selection and drill down through several layers of information to choose records that met a specific criterion, was of great interest to the participants. Also, learning how to subset data, edit record entries (e.g., coordinates) using QGIS, and save the cleaned and appropriately georeferenced records as comma-delimited (or comma-separated) text files (CSV) was of great interest and added to the tutorial.
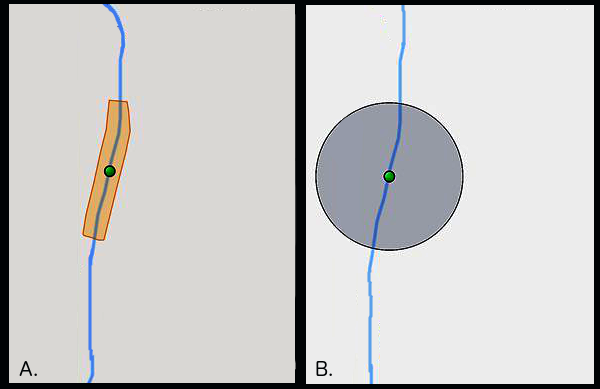
Georeferencing among data managers and researchers in biocollections generally use the point-radius method (Fig.
An illustrative example of the two methods of uncertainty capture when georeferencing specimens. Method A, or polygon, creates a shape around the river (in blue). Method B, or point-radius, creates a circle of uncertainty around the origin. The illustration is based on output from GeoLocate software (
Researchers and data managers were taught how to import existing georeferenced collections into software such as QGIS. Once spatially referenced, it is possible to couple the georeferenced data with existing layers of spatial information that can supplement assessment and analysis. Some layers of interest included administrative boundaries (for checking centroids, or center points of polygons, against existing records), ecoregions, cultivated gardens and zoos, and parks and protected areas. Other researchers expressed interest in acquiring and incorporating data about elevation, federally managed sites, and climate data. A conversation about where to find data resources, like local municipality GIS office websites and ArcGIS Online’s Living Atlas of the World, proved helpful.
On the first day of the QGIS tutorial, the topics covered included importing tutorial data obtained from iDigBio, adding additional layers, and saving a map project. The following day included advanced topics, such as performing a spatial join across layers to query attributes, sub-setting the dataset based on spatial selections and intersection, changing the symbology of the data, and performing summary statistics on the attributes. Additionally, visualizing the data over time and producing heat maps of observation locations provided new views on trends within the dataset that would be impossible to detect by simply viewing the records in a table.
Reactions from the data managers and researchers about the QGIS tutorial was positive. Many were excited about the prospect of incorporating QGIS and other spatial analysis techniques into their georeferencing workflows. While fewer participants indicated interest in using QGIS for data cleaning, many expressed interest in using the techniques for data exploration and public communication. Participants who brought their own datasets to the workshop spent time on the last day running similar analyses against their collections in QGIS and were interested in learning how to perform specific GIS operations to spatialize their research questions. For example, several participants were interested in assessing the spatial distribution of specimens in their study area to find a region with a greater significant number occurences. Others were interested in testing predictions about regions where the specimens were not likely to occur (e.g., studying absence) and using other spatial information to account for the trends in distribution.
Desired skills to develop included coupling QGIS with Python scripting to enable researchers and data managers to work with larger datasets. Researchers who also wanted to incorporate QGIS into georeferencing workflows were interested in plugins such as QGIS Gazetteer Plugin that would allow them to cross-reference observations and flag records for removal from further analysis. Best practices for editing CSV files in QGIS were also desired, and a lack of current consensus surrounding when to remove a point from a dataset resulted in inconsistent practices surrounding editing of data using QGIS. For example, once a point has been flagged because it corresponds too closely to a county centroid, should it be removed from the dataset or can its position be rectified? Should the record be noted but not included in analysis? Ideally, the modified and annotated research dataset would be published with original identifiers to enable linking of the data to the original data record and the physical specimen.
Many researchers and collections managers expressed interest in a follow-up tutorial on more advanced techniques in QGIS and were highly motivated to incorporate QGIS into their workflows upon completion of the tutorial. Interest in using QGIS for georeferencing tasks, data exploration, and public communication indicated that planned future tutorials could help users apply more advanced techniques to their existing research or collections datasets.
Participant surveys and evaluations
We gathered data from the participants using pre, post and follow-up surveys. Results from these were used to gauge enthusiasm from participants for the workshop and the direction participants thought future training should progress. The pre-workshop data was collected online through the workshop application using a google forms (Suppl. material
Pre- and post-workshop surveys
Post workshop surveys were given to participants at the end of each day. All participants recorded their knowledge of georeferencing best practices and resources, and their skills for creating and using spatial data were higher after the workshop, with 63-68% of participants indicating a much higher knowledge level after the workshop.
The post-workshop surveys show that more than 50% of participants rated all but two topics covered in the workshop as "most valued." Comments on the survey suggest that the two, less-highly rated topics Good-Bad Localities and Getting Datasets, were seen as review rather than introduction of new information. Participants also responded that the most useful topics provided during this workshop were OpenRefine and QGIS, the advanced features of GEOLocate, and how to incorperate available APIs into workflows for both basic georeferencing and research.
Table
Perceived community needs for tools, standards, and skills and training needs.
|
High |
Somewhat High |
Neither High nor Low |
Somewhat low |
|
|
Improving georeferencing efficiency (tools, training) |
11 |
7 |
1 |
|
|
Georeferenced data sharing and reintegration |
10 |
7 |
2 |
|
|
Quality or fitness-for-use indicators for georeferenced data (standards) |
9 |
7 |
2 |
1 |
|
Visualizing georeferenced data (training) |
9 |
8 |
1 |
1 |
|
QGIS for spatial analysis (training) |
8 |
7 |
4 |
|
|
R scripting (training) |
8 |
5 |
5 |
1 |
|
GEOLocate (training) |
7 |
7 |
3 |
2 |
|
OpenRefine (training) |
7 |
9 |
3 |
|
|
Developing georeferencing expertise (training) |
7 |
7 |
5 |
|
|
Darwin core georeferencing fields (standards) |
4 |
10 |
5 |
|
|
Gazetteer development/ availability (tools. standards) |
3 |
12 |
2 |
2 |
Follow-up survey
Fifty-nine percent of the participants responded to the follow-up survey distributed three months following the workshop (Suppl. material
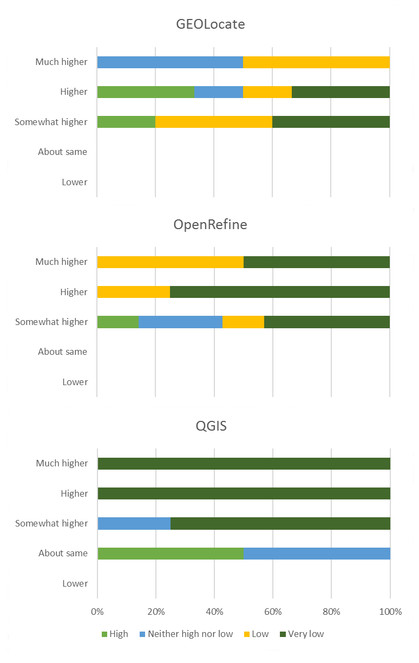
Initial expertise (color of the bar) vs final confidence (y-axis) after the GRU workshop for participants responding to final survey. Example for how to interpret this graphic: the blue color bar at the top indicates that before the workshop roughly 50% of respondents said their knowledge of GEOLocate was "neither high nor low" but after the workshop these same respondents selected "much higher" for their knowledge of GEOLocate.
Topics for future workshops
A complete list of topics and new tools that participants identified during the workshop as something they wanted to learn more about is available in the supplementary files (Suppl. material
Discussion
Digitization of biocollections is somewhat unique in the biological and paleontological data community because the major funding sources that create the data often do not include financial support for research using the resulting digitized data products. This produces large-scale data capture projects that do not participate in research efforts using those data, and these data providers (both collections and aggregators) need input from data users to improve their data products.
One of the expectations between data providers and data users involves the quality of the georeferenced coordinates. Data providers have a mandate to deliver data efficiently, and often data users require data with a quality than may not be cost-efficient or feasible to produce. Other times, locations are deliberately obfuscated (see dwc:informationWithheld) in public databases due to biosecurity, privacy, sensitive species, and other legal reasons. Original locations are sometimes stored, and the less-precise georeferences are displayed. No currently existing methods that we know of offer easy ways to ensure these withheld data get published when finally appropriate to do so, if ever.
The strongest finding was that 62% of workshop participants increased their use of QGIS since the workshop and 77% expect to use the software increasingly over time, highlighting the importance of training opportunities for career professionals in all sectors of the biodiversity data realm. Expressions of interest by participants for future training on other topics of interest related to georeferencing and the use of biocollections data have been outlined in the iDigBio GRU Wiki requests for the future, and in the post-workshop survey.
Biocollection workflows are changing
Key conversations at the workshop centered around capturing historical versus future biocollections data. All participants, data researchers and data providers noted changes occurring in biocollections data workflows, yet outstanding challenges continue. For all of these reasons and more, there exists a need for ongoing initiatives to address these topics.
- Significant quantities of specimen data collected remain on paper specimen labels, in notebooks, catalogs, and field cards, limiting online open access and resulting in patchy datasets.
- At the same time, new specimens are still being accessioned into biocollections in solely paper-based data formats, thus further contributing to a largely inaccessible backlog of biodiversity specimen data.
- Backlogged legacy data is labor-intensive, expensive, and sometimes difficult to georeference, and often has significant uncertainty.
- Not all georeferences (new or legacy) come with the metadata needed for researchers (or algorithms) to effectively evaluate geospatial data fitness-for-use.
- Many collections undertake the time-consuming process to provide uncertainty data using point-radius or polygons, however, these data seem to be underused in scientific analysis.
Collections and researchers need workflows that reduce unnecessary future collections data management and speed access to data. On this topic, our discussions focused on best practices for creating new georeferences when collecting specimens, georeferencing existing collection locality data, and methods for use and evaluation of geospatial coordinates from historic biocollections records. Optimistically, technology is making it possible for new specimen data to be born-digital (
Importance of access to centralized resources
Finding materials and expertise can be challenging and time-consuming. Development of new tools and workflows continues. Participants appreciated consolidated resources like those found at iDigBio and the Biodiversity Catalogue. Aggregation of this information saves time and effort. Some topics and tools covered stood out from the rest for all participants. These include: template generation for data collection, such as the Biocode Field Information Management System, application development with Open Data Kit (mobile-first data collection platforms), and discussion and development of a data validation process (Suppl. material
Cross-discipline interactions
Hands-on interaction with new software tools and APIs for data refinement, along with in-person interactions enabled by the workshop improved the efficiency of the learning process. Sharing standards of practice and needs across disciplines highlighted the need for changes and transparency in future data collection, evaluation, and data sharing processes.
Exploring and encouraging use of uncertainty
Our workshop experience suggests the need to improve creation, use, and publication of uncertainty data. Whether expressed as a polygon or point-radius, this information appears to be underused and quite time-intensive to create. The biocollections and research communities are encouraged to utilize best practices for creating and sharing coordinate uncertainty information, and the collections community need research examples using these data in order to justify such effort. This may require future training in the research use and value of uncertainty data.
Improved communication about data quality
Data aggregators like GBIF, Atlas of Living Australia, and iDigBio are working through the Biodiversity Science and Standards TDWG/GBIF Biodiversity Data Quality Interest Group to harmonize biocollections data quality (DQ) feedback. Aggregators provide these standardized DQ assertions: 1) to those browsing the data online, 2) to the data providers, and 3) in any downloaded datasets. However, this DQ process, along with the tools, methods, and data standards used during data capture and publishing are not always understood in the research community. For instance, data aggregators currently use taxonomic name assemblages to facilitate searching and indexing of aggregated data. But as a result, the taxonomy may not be what some expect or need (
Proposed actions to speed up biocollection data in research
Some possible actions to advance and speed up the use of biocollection data in research are listed here, and the worldwide biodiversity collections network, industry, and groups like the iDigBio GWG are encouraged to collaborate to support:
- increased outreach and training of necessary skills, standards, and literacy in the biodiversity data community,
- georeferencing of specimens at the time of collection (including uncertainty and source of the coordinates),
- further development and integration of tools such as GEOLocate into collection management systems,
- streamlining batch processing (
Guo et al. 2008 ), - the development of shared locality services and more gazetteer resources to reduce repeated georeferencing efforts and improve usefulness, and
- the development of techniques to publish and link georeferenced research data sets to the original occurrence records and physical specimens.
Conclusions
The Georeferencing for Research Use workshop was a successful workshop based on the discussion, survey results, and issues reflected in the captured conversations. It created an important platform for biocollections data providers to learn directly from the researchers who hope to use the data they provide and vice versa. As we continue to provide data about specimens, and learn to use the data in research, workshops that provide this kind of cross talk will continue to be important learning platforms that will improve the quality of research and data products. Workshops of this type also offer strategic opportunities to discover future leaders and innovators in our community as the role of collection and data managers evolve to support faster data mobilization and more robustly standardized and complete datasets. We anticipate data collated and summarized in this survey report will contribute valuable information for planning future activities.
Funding program
iDigBio is funded by grants from the US National Science Foundation's Advancing Digitization of Biodiversity Collections program (Co-operative Agreements EF-1115210 and DBI-1547229). Additional funding was provided by the Cheadle Center for Biodiversity and Ecological Restoration, University of California, Santa Barbara.
Training was also made possible due to the following US National Science Foundation funded programs: GEOLocate (DBI-1202953, DBI-0852141, DBI-0516312 and DBI-0131053), VertNet (DBI-062148), HerpNET (DBI-0108161), FishNet2 (DBI-0417001), ORNIS (DBI-0345448), and MaNIS (DBI-0108161).
Hosting institution
University of California, Santa Barbara National Center for Ecological Analysis and Synthesis and University of California, Santa Barbara Cheadle Center for Biodiversity and Ecological Restoration.
Ethics and security
The application form, post-workshop surveys, and follow-up survey questions and protocols were reviewed and approved by the University of Florida Human Subjects Committee Institutional Review Board (IRB) (University of Florida IRB201601849).
Author contributions
Deborah L. Paul and Katja C. Seltmann conceptualized the workshop; Deborah L. Paul, David Bloom, Nelson Rios, Shelley A. James, Sara Lafia, Shari Ellis, Katja C. Seltmann, Una Farrell, Jessica Utrup and Michael Yost developed the workshop teaching materials; Edward Davis, Rob Emery, Gary Motz, Julien Kimmig, Vaughn Shirey, Emily Sandall, Daniel Park, Christopher Tyrrell, R. Sean Thackurdeen, Matthew Collins, Vincent O'Leary, Heather Prestridge, Christopher Evelyn, and Ben Nyberg participated in the workshop and contributed to the manuscript.
References
-
Final Report of the Task Group on GBIF Data Fitness for Use in Distribution Modelling.GBIF Secretariat,Copenhagen. URL: http://www.gbif.org/system/files_force/gbif_resource/resource-82612/Final_report_of_the_task_group_on_GBIF_data_fitness_for_use_in_distribution_modelling_ver1.1.pdf?download=1
-
Best Practice Guide for Data Gap Analysis for Biodiversity Stakeholders. https://www.gbif.org/document/82566/best-practice-guide-for-data-gap-analysis-for-biodiversity-stakeholders. Accessed on: 2018-5-18.
-
Final report of the Task Group on GBIF data fitness for use in agrobiodiversity.GBIF Secretariat,Copenhagen. URL: https://www.gbif.org/document/82283/report-of-the-task-group-on-gbif-data-fitness-for-use-in-agrobiodiversity
-
Emerging technologies for biological recording.Biological Journal of the Linnean Society115(3):731‑749. https://doi.org/10.1111/bij.12534
-
Why georeferencing matters: Introducing a practical protocol to prepare species occurrence records for spatial analysis.Ecology and Evolution8:765‑777. https://doi.org/10.1002/ece3.3516
-
Green plants in the red: A baseline global assessment for the IUCN sampled Red List Index for plants.PLOS One10(8):e0135152. https://doi.org/10.1371/journal.pone.0135152
-
Guide to best practices for georeferencing.Global Biodiversity Information Facility,Copenhagen, Denmark,28pp. URL: http://www.gbif.org/orc/?doc_id=1288
-
Biocode Field Information Management System. http://www.biscicol.org/. Accessed on: 2017-3-03.
-
What is Biosecurity?https://www.agric.wa.gov.au/biosecurity-quarantine/biosecurity. Accessed on: 2018-9-23.
-
To increase trust, change the social design behind aggregated biodiversity data.Database: The Journal of Biological Databases and Curation2018(1):bax100. https://doi.org/10.1093/database/bax100
-
Georeferencing locality descriptions and computing associated uncertainty using a probabilistic approach.International Journal of Geographical Information Science22(10):1067‑1090. https://doi.org/10.1080/13658810701851420
-
Big data and the future of ecology.Frontiers in Ecology and the Environment11(3):156‑162. https://doi.org/10.1890/120103
-
Skills and knowledge for data-intensive environmental research.BioScience67(6):546‑557. https://doi.org/10.1093/biosci/bix025
-
Biodiversity online: Toward a network integrated biocollections alliance.BioScience63(10):789‑790. https://doi.org/10.1525/bio.2013.63.10.4
-
Why Digitize?https://vimeo.com/184552432. Accessed on: 2016-1-01.
-
iDigBio Portal Search. www.idigbio.org/portal/. Accessed on: 2018-9-23.
-
microclim: Global estimates of hourly microclimate based on long-term monthly climate averages.Scientific Data1https://doi.org/10.1038/sdata.2014.6
-
Accelerating the discovery of biocollections data. http://www.gbif.org/resource/83022. Accessed on: 2018-4-04.
-
Biodiversity analysis in the digital era.Philosophical Transactions of the Royal Society B: Biological Sciences371(1702):20150337. https://doi.org/10.1098/rstb.2015.0337
-
Museums, Paleontology, and a Biodiversity Science Based Approach. In: Rosenberg GD, Clary RM (Eds)Museums at the Forefront of the History and Philosophy of Geology: History Made, History in the Making.Vol. 535.Geological Society of Americahttps://doi.org/10.1130/2018.2535(22)
-
Natural history collections as sources of long-term datasets.Trends in Ecology & Evolution26(4):153‑154. https://doi.org/10.1016/j.tree.2010.12.009
-
Estimating species diversity and distribution in the era of Big Data: to what extent can we trust public databases?Global Ecology and Biogeography24(8):973‑984. https://doi.org/10.1111/geb.12326
-
Harnessing the power of Big Data in biological research.BioScience63(9):715‑716. https://doi.org/10.1525/bio.2013.63.9.4
-
Task group on data fitness for use in research into invasive alien species.GBIF Secretariat,Copenhagen. URL: https://www.gbif-uat.org/document/82958/data-fitness-for-use-in-research-on-alien-and-invasive-species
-
An audit of some processing effects in aggregated occurrence records.ZooKeys751:129‑146. https://doi.org/10.3897/zookeys.751.24791
-
Equipping the 22nd-century historical ecologist.Trends in Ecology & Evolution32(8):578‑588. https://doi.org/10.1016/j.tree.2017.05.006
-
A place for everything.Nature453(7191):2‑2. https://doi.org/10.1038/453002a
-
OCLC Research: Demystifying Born Digital. https://www.oclc.org/research/themes/research-collections/borndigital.html. Accessed on: 2018-3-19.
-
When natural history collections reveal secrets on data deficient threatened species: Atlantic seahorses as a case study.Biodiversity and Conservation26(12):2791‑2802. https://doi.org/10.1007/s10531-017-1385-x
-
Implications and alternatives of assigning climate data to geographical centroids.Journal of Biogeography44(10):2188‑2198. https://doi.org/10.1111/jbi.13029
-
GEOLocate - Software for Georeferencing Natural History Data. [Web application software].http://www.geo-locate.org. Accessed on: 2018-4-09.
-
LepNet: The Lepidoptera of North America Network.Zootaxa4247(1):73‑77. https://doi.org/10.11646/zootaxa.4247.1.10
-
The role of natural history collections in documenting species declines.Trends in Ecology & Evolution13(1):27‑30. https://doi.org/10.1016/s0169-5347(97)01177-4
-
SimpleMappr, an online tool to produce publication-quality point maps.Accessed December4:2018. URL: http://www.simplemappr.net.
-
Erosion of Lizard Diversity by Climate Change and Altered Thermal Niches.Science328(5980):894‑899. https://doi.org/10.1126/science.1184695
-
Changes in data Sharing and data reuse practices and perceptions among scientists worldwide.PLOS One10(8):e0134826. https://doi.org/10.1371/journal.pone.0134826
-
Data from drawers: Securing, mobilising and interrogating National Research Collections data. In: National Collections Research Australia (NCRA)Digitization of Biological Collections: A Global Focus.CSIRO (Commonwealth Scientific and Industrial Research Organisation), Blackmountain Labs, Canberra,April 2015.
-
New knowledge from old data: the role of standards in the sharing and reuse of ecological data.Science, Technology, and Human Values33(5):631‑652. https://doi.org/10.1177/0162243907306704
Supplementary materials
Darwin Core Archive file downloaded from the iDigBio portal for use in the Georeferencing for Research Use workshop. Total 25,429 records, accessed on 2016-08-29. Collections contributing to the record set are listed in the archive records.citation.txt file. Dataset GUID: a69d1541-4726-465d-84ad-50c7ed556eee
Download file (5.67 MB)
This document shows just the questions we asked the applicants who applied to participate in this Georeferencing for Research Use workshop. We used a Google Form to deliver these questions and collect responses. It is both an application and serves as our pre-workshop survey.
Download file (39.64 kb)
The informed consent request and workshop survey questions given to participants after the workshop each day for 4 consecutive days.
Download file (178.88 kb)
Three months after the workshop, participants were surveyed to assess what workshop-related knowledge and materials were being used and disseminated to others. This document summarized data collected in this particular survey.
Download file (225.30 kb)
Questions we asked in the Georeferencing for Research Follow Up Survey done 3 months after the workshop.
Download file (67.08 kb)
Summary of topics to be covered in an ideal workshop as identified by workshop applicants in the workshop call for participation. We incorporated as many as possible that also fit our scope.
Download file (8.38 kb)
This document contains an annotated set of data quality checks that participants report they use when evaluating and cleaning datasets. These items outline how participants are judging if the data suits their purpose.
Download file (30.67 kb)
Summary of desired future workshop topics that were listed by participants on the last day of the workshop.
Download file (13.33 kb)