|
Research Ideas and Outcomes :
Grant Proposal
|
|
Corresponding author: Lyubomir Penev (l.penev@pensoft.net)
Received: 25 Jan 2022 | Published: 26 Jan 2022
© 2022 Lyubomir Penev, Dimitrios Koureas, Quentin Groom, Jerry Lanfear, Donat Agosti, Ana Casino, Joe Miller, Christos Arvanitidis, Guy Cochrane, Donald Hobern, Olaf Banki, Wouter Addink, Urmas Kõljalg, Kyle Copas, Patricia Mergen, Anton Güntsch, Laurence Benichou, Jose Benito Gonzalez Lopez, Patrick Ruch, Corinne Martin, Boris Barov, Iliyana Demirova, Kristina Hristova
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Penev L, Koureas D, Groom Q, Lanfear J, Agosti D, Casino A, Miller J, Arvanitidis C, Cochrane G, Hobern D, Banki O, Addink W, Kõljalg U, Copas K, Mergen P, Güntsch A, Benichou L, Benito Gonzalez Lopez J, Ruch P, Martin CS, Barov B, Demirova I, Hristova K (2022) Biodiversity Community Integrated Knowledge Library (BiCIKL). Research Ideas and Outcomes 8: e81136. https://doi.org/10.3897/rio.8.e81136
|
|
Abstract
BiCIKL is an European Union Horizon 2020 project that will initiate and build a new European starting community of key research infrastructures, establishing open science practices in the domain of biodiversity through provision of access to data, associated tools and services at each separate stage of and along the entire research cycle. BiCIKL will provide new methods and workflows for an integrated access to harvesting, liberating, linking, accessing and re-using of subarticle-level data (specimens, material citations, samples, sequences, taxonomic names, taxonomic treatments, figures, tables) extracted from literature. BiCIKL will provide for the first time access and tools for seamless linking and usage tracking of data along the line: specimens > sequences > species > analytics > publications > biodiversity knowledge graph > re-use.
Keywords
biodiversity data, data access, research cycle, data linking, FAIR data, identifiers
List of participants
Detailed list of all paricipants is available in Table
| List of participants | |||
|---|---|---|---|
| Participant No | Participant organisation name | Abbreviation | Country |
| 1 (Coordinator) | Pensoft Publishers | PENSOFT | Bulgaria |
| 2 | Naturalis Biodiversity Center | NATURALIS | The Netherlands |
| 3 | Plazi GmbH | Plazi | Switzerland |
| 4 | Meise Botanic Garden | MeiseBG | Belgium |
| 5 | European Molecular Biology Laboratory | ELIXIR/EMBL-EBI | IEOI |
| 6 | European Organization for Nuclear Research | CERN | Switzerland |
| 7 | Consortium of European Taxonomic Facilities | CETAF | Belgium |
| 8 | Swiss Institute of Bioinformatics | SIB | Switzerland |
| 9 | University of Tartu | UTARTU | Estonia |
| 10 | LifeWatch ERIC | LIFEWATCH | Spain |
| 11 | Freie Universitaet Berlin | FUB-BGBM) | Germany |
| 12 | Global Biodiversity Information Facility | GBIF | Denmark |
| 13 | Species 2000 | Sp2000 | The Netherlands |
| 14 | Stichting International Working Group on Taxonomic Databases | TDWG | The Netherlands |
Third parties involved in the project
Muséum Nationale d’Histoire Naturelle (MNHN), Paris will act as a linked third party to CETAF.
Excellence
Objectives
Summary
BiCIKL will initiate and build a new European starting community of key research infrastructures in biodiversity and life sciences, solidifying open science practices through provision of access to data, associated tools and services at
- each separate stage of, and
- along the entire research cycle.
BiCIKL will provide for the first time seamless access, linking and usage tracking of data within a network of links between the different data classes, ulti- mately represented in the biodiversity knowledge graph: specimens → genetic sequences → species → analytics → publications → biodiversity knowledge graph → re-use. BiCIKL will also provide new methods and workflows for an integrated access to harvesting, liberating, linking, and re-using of sub-article-level data (specimens, material citations, samples, sequences, taxonomic names, taxonomic treatments, figures, tables) extracted from literature.
The added value of the new community over the sum of the existing services, besides the improved access at each stage of the data and research life cycle, will be the provision of a single knowledge broker, the Biodiversity Knowledge Hub (BKH), to interlinked, machine-readable, Findable, Accessible, Interoperable and Reusable (FAIR) data connecting specimens, genomics, observations, taxonomy and publications. The existing services provided by the participating infrastructures will be expanded through the development and adoption of shared/common/interoperable domain standards which will liberate and enhance the flows of data and knowledge across these domains. Looking forward and through incorporating lessons learned from the joint research activities and feedback from the access provided to researchers, BiCIKL will make possible the establishment of next-generation scholarly practices entirely based on open data and open science principles. The novel tools and workflows developed for extraction, FAIRification, management and re-use of data extracted from literature, and those that provide prospective, data- and narrative-integrating publishing, can be used in its generic form in domains beyond biodiversity. While respecting the European Open Science Cloud (EOSC) principle “Data as open as possible, as closed as necessary”, BiCIKL will focus on data that are already Open or such that will be made Open and FAIR during the project and beyond it.
The project will focus on the following overall objectives integrated across three main pillars (Access, Networking, Joint Research Activities):
- Find: Ensure seamless discoverability of data through globally unique identifiers exposed to individual and federated search engines, including artificial intelligence, from each participating infrastructure and across data domains.
- Access: Provide, facilitate, support and scale up open access to FAIR interlinked data, liberated from literature, natural history collections, sequence archives and taxonomic nomenclators in both human-readable and machine-actionable formats.
- Interoperate: Harmonise the existing standards, metadata, policies and technologies and develop new ones, where necessary, for provision and ingestion of FAIR data to ensure standard-aligned interlinking and re-use between data domains.
- Re-use: Optimise the reusability and reproducibility of complex datasets, assembled together from different biodiversity-related domains and their supporting infrastructures, for generation of research hypotheses and new knowledge.
The key products of this project will be:
- A vibrant community equipped with novel research tools for search and access to data interlinked across domains.
- Interlinked corpora of knowledge used by research groups in biodiversity science and related areas.
- Automated text and data mining workflows for extraction, XML and RDF conversion, semantic enhancement, management, dissemination, and re-use of the huge amount of highly valuable data linked to the Linnean names of species, accumulated in the legacy literature.
- Semantics-based journal production workflows for the community but also as a seed for adoption by other communities.
Rationale
The science of biodiversity has accumulated probably one of the oldest and richest data pools on the living world, dating back to the ancient times and resulting in more than 500 million pages of published literature, more than 2 billion specimens in natural history collections and more than 1.8 million species described (
In parallel to this deluge of raw data (especially in light of the decline in sequencing costs), scholarly publications comprise knowledge based on billions of facts in millions of published narratives. This corpus of knowledge actually represents a rich citation network, albeit almost entirely based on implicit citation links. These links set by the authorities (authors) have the potential to be a base for building a biodiversity knowledge graph (
On the other side, some digitally born publications in the domain of biodiversity provide explicit links, embedded during the act of publishing, for example to DNA sequences, digital natural history specimens, species identifications, literature citations, people and ontologies (
Despite these positive developments, the larger these corpora of knowledge become, the larger is the disconnection gap between them (
- specimens and samples,
- genomics,
- taxon names and OTUs,
- literature, and
- analytics.
The starting community will provide a new level of access to FAIRified data from (A) each separate domain-specific infrastructure, (B) bi- and/or multi-directionally linked data across domains, including unprecedented access to (C) data currently imprisoned in millions of pages of published literature. In order to mobilise new research and data from prospectively published literature and make it immediately accessible and re-usable in the research cycle, BiCIKL will expand and promote (D) next-generation semantic publishing workflows which will provide immediate access to the data therein and thus remove the high cost of data liberation for the users of the future.
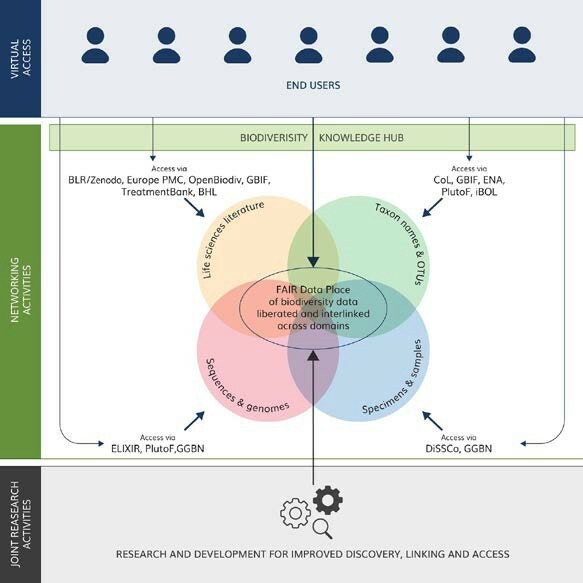
The rapidly developing landscape of European and international research data infrastructures has worked to ensure that data becomes more FAIR and that services are developed to enable users to discover, access and use types of data deposited across a multitude of infrastructures (GBIF, ENA, BLR, DiSSCo and others) (
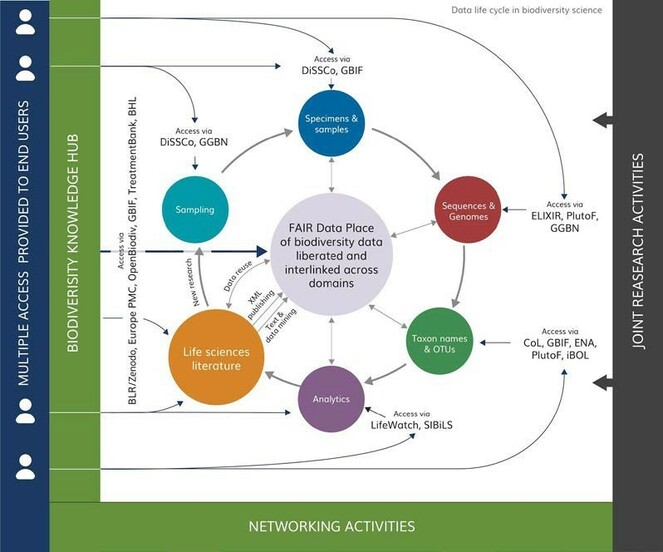
The Biodiversity Knowledge Hub (BKH), the main product of BiCIKL, will act as a knowledge broker for unparalleled access to data and associated tools and services made available through integration of state-of-the-art research infrastructures. Through integration and synergies of existing research infrastructures, BiCIKL will provide a completely new level of services at the disposal of the researchers, including enabled access to FAIR data at any stage of the research cycle. These services will also open the future generation approach to harvesting, liberating, accessing and re-using data from the biodiversity and life sciences literature, from specimen and sample collections, from genomic sequences and taxonomic units, for anybody, anytime and from anywhere.
In summary, as illustrated in Fig.
Specific objectives
To accomplish its key objectives to provide access to the interlinked corpora of knowledge, BiCIKL will:
- Develop and implement open science research practices for provision of multi-modal Trans-national (TA) and Virtual (VA) access to Research Infrastructures (RIs), data and tools along the entire research cycle in the field of biodiversity and related areas. (Measured by the number of units of access in TA and various usage metrics (number of users and uses (e.g., data downloads, unique and total visits) in VA).
- Harmonise policies, standards and technologies for access to data and services between the participating key infrastructures. (Measured by published standards regulating bi- and multi-directional linking and access between RIs).
- Engage all actors and other stakeholders in the process of access and use of data on both ends of the spectrum (data upload/ingestion and FAIR data delivery): research infrastructures, researchers, industry partners, public bodies and other end users. (Measured by the number of requests for and uses of bi- and multi-directional linking and access).
- Build and improve researchers’ capacity through enhanced digital skills and competence in uptaking new levels and methods of access and re-use of linked open data. (Measured by the number of access to/downloads of guidelines and by the number of participants in training events).
- Provide a one-stop access point to guidelines, standards, data and services via the newly developed Biodiversity Knowledge Hub (BKH), hosted and maintained by a large international organisation (GBIF). (Measured by the number of visits to the BKH website and followers of the BKH social media profiles).
- Foster joint research agendas of European and international researchers through open call projects demon trating the value of access to multiple data sources across different domains. (Measured by the number of applications for open call projects).
- Support industrial innovation provided by the participating SMEs in building and implementation of (1) next-generation, standards-aligned and semantics-based publishing workflows and (2) novel automated workflows for mining, liberation and FAIR-fication of data from the literature. (Measured by the number of users of the enhanced publishing and data mining tools and services, and by the number of journals and publishers that adopted the novel publishing workflows).
- Liberate and re-use the vast knowledge and data imprisoned in the published narratives for centuries of biodiversity research. (Measured by the numbers of processed articles and pages, and extracted sub-article data elements (taxon treatments, accession numbers, figures, material citations, tables and others).
- Support researchers’ access to the big Linked Open Data world through interoperable, AI-based, FAIR Data Place (FDP) interface, discovering and validating links between data from different resources. (Measured by the number of access to the FDP endpoint).
- Facilitate inter-disciplinary research cooperation and generation of new knowledge through enhanced and newly developed tools for interlinking of FAIR data from different resources and domains. (Measured by manuscripts, submitted as a result of the research projects using interlinked FAIR data).
Relation to the work programme
The INFRAIA-02 call focuses on integration on European scale, and opening up key research infrastructures to all European researchers, from both academia and industry, ensuring their optimal use and joint development.
BiCIKL will build a new European ‘starting community’ involving the 16 leading research infrastructures of European and global interest in the Biodiversity and Life Science domains. The main product of the integration of these infrastructures will be the Biodiversity Knowledge Hub (BKH). The users of BKH will receive unprecedented access to data, tools and services currently only partly accessible via scattered locations on the web. Moreover, by using advanced text and data mining tools, BiCIKL will open for the first time a vast corpus of inaccessible data and knowledge ‘imprisoned’ in publications.
BiCIKL response to the general objectives and aims of the programme
More specifically, the response of BiCIKL to the general policy requirements of the work programme are listed in Table
BiCIKL response to the key requirements of the INFRAIA-02-2020 programme.
|
|
Item |
Call text |
Project contribution / gaps |
Location in proposal |
|
1 |
Overall aim of the Work Programme 2018-2020 |
The Research Infrastructures Work Programme 2018-2020 contributes to the implementation of the ESFRI Roadmap. Fostering long-term sustainability of RI and expanding their role and impact. |
BiCIKL will advance the ESFRI (European Strategy Forum on Research Infrastructures) Roadmap by clustering of Research Infrastructures and improving their horizontal linkages, and the adoption of the Open Science concept, in full alignment with Recommendation 3 of ESFRI’s Long-term Sustainability Working Group* BiCIKL will lay the ground for a novel pan-European ecosystem of well-established, but currently not well-connected RIs. Such integration will be carried out at bi- and multi-directional levels across RIs to avoid duplication of effort and provide improved access to interlinked FAIR data for researchers, public authorities, business and any user, including the general public. |
Sections "BiCIKL contributing to the ESFRI Roadmap" (Fig. |
|
2 |
Policy objectives |
Priority 1 of the Juncker Commission: A Connected Digital Single Market, to open Big Data to researchers, innovators and business. |
BiCIKL will connect data from different domains, adding to them a huge corpus of data, including such liberated from literature into a big FAIR data pool seamlessly available to researchers, public authorities and business to foster innovation in the digital economy.
|
Sections "BiCIKL contributing to the ESFRI Roadmap", "Improved access via BiCIKL as a basis for cross-cut research", "Innovation is the key for BiCIKL to establishing a successful new community", "The BiCIKL community in a ten-years perspective"; NA-01-03, JRA-01,JRA-05; Outcomes D1.3, D11.3, D11.5. |
|
|
Policy objectives |
Priority 1 of the v.d. Leuven Commission: A Green European Deal.
|
One set of actions under the European Green Deal relates to protecting/restoring biodiversity. All data provided in a novel format and usage potential will serve as a background evidence to improve policies and practices of nature conservationists and practitioners towards preserving Europe’s and World’s natural heritage. |
Sections "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "Improved access via BiCIKL as a basis for cross-cut research"; NA-02-03; JRA-10, TA, VA; Outcomes D3.3-3.4, D4.4. D5.1. |
|
|
Policy objectives |
The use of European Structural and Investment Funds to build capacities and infrastructures at national and regional level. |
One of the BiCIKL partners, Pensoft, used structural funds (Competitiveness) in 2008-2010 to create its first journal publishing platform TRIADA, which laid the ground for the subsequent successful innovation and growth of the company. The contribution of Pensoft to BiCIKL builds on this prior public investment. |
Section "Innovation is the key for BiCIKL to establishing a successful new community"; JRA-01, TA; Outcomes D6.1, D6.4. |
|
|
Policy objectives |
Support to actions included in the 2016 Communication on the European Cloud Initiative, in particular to further integrate and consolidate e-infrastructure platforms, to connect the ESFRI infrastructures |
The European Cloud Initiative aims to unlock the power of Big Data for open science and innovation. This is particularly true for biodiversity knowledge which has traditionally lagged behind in contrast to e.g. “harder” sciences such as remote sensing and earth observation more generally. Furthermore, BiCIKL comes at a time when the decreasing cost of sequencing and novel methods such as environmental DNA (eDNA) will generate enormous amounts of data. As a result of BiCIKL, data from across |
Sections "BiCIKL contributing to the ESFRI Roadmap", "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "Innovation is the key for BiCIKL to establishing a successful new community", "Strengthening the European Research Area (ERA): Europe as a global leader in open data"; JRA-06-10, TA, VA; Outcomes D4.3-4.4, D5.1, D6.4, D11.5. |
|
|
|
to the European Open Science Cloud, and to develop a European Data Infrastructure. |
domains (literature, images, sequences, specimens, taxon names, taxonomic trees) and other corpora of knowledge will be presented as Linked Open Data (LOD) and biodiversity knowledge graph, which can be openly used and interlinked with any other LOD across the European Open Science Cloud and beyond, at global-scale. |
|
|
|
Policy objectives |
Strategy for EU international cooperation in research and innovation (COM(2012)497).
|
Research infrastructures typically rely on large investments by public and charitable funders for their establishment and operation. This is usually made possible through international cooperations, as illustrated by two project partners EMBL (with EMBL-EBI and ELIXIR Hub) and CERN, which are intergovernmental organisations of European interest. Furthermore, ELIXIR and CERN have been recognised as RIs of Global Interest by the Group of Senior Officials on Global Research Infrastructures of the G7* |
Sections "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "The BiCIKL contribution to fostering a culture of international cooperation", "Innovation is the key for BiCIKL to establishing a successful new community", "The BiCIKL community in a ten-years perspective", "Strengthening the European Research Area (ERA): Europe as a global leader in open data"; NA-03, JRA-01-05, TA, VA; Outcomes D3.3-3.4, D4.3-4.4, D5.1. |
|
3 |
Data sharing policy |
Grant beneficiaries under this work programme part will engage in research data sharing by default, as stipulated under Article 29.3 of the Horizon 2020 Model Grant Agreement and Data Management Plan. |
Open science is at the core of the organisational missions of all BiCIKL partners. All data that are subject of work in BiCIKL will hence be available as FAIR data under CC0 (public domain) or CC-BY license, according to a DMP elaborated and agreed upon in the project. The Bouchout Declaration of Open Biodiversity Knowledge Management, launched by the EU funded pro-iBiosphere project, including BiCIKL partners (Plazi, Pensoft, MBG, Naturalis, BGBM-FUB), and endorsed by CETAF and 96 organizations worldwide, will be used as a conceptual basis of the policy framework of BiCIKL. |
Sections "BiCIKL as an entirely open science project, from start to end", "Dissemination and exploitation of results", "Policies, processes and standards harmonisation across organisations"; NA-01-03, JRA-01-06, VA, TA; Outcomes D3.4, D4.3-4.4, D5.1, D6.3-6.4, D11.5. |
|
4 |
Aims of the actionSpecific aim |
To bring together, integrate on European scale, and open up key national and regional research infrastructures to all European researchers, from both academia and industry, ensuring their optimal use and joint development. |
Most of the key partners have sound experience in coordinating RIs at global (GBIF), European (DiSSCo, ELIXIR, SYNTHESIS+) and national level (details in Sect. 3.3.1). More specifically, BiCIKL will build a new inter-disciplinary starting community that enables scientists, policy makers and industrial partners to access the complex corpora of knowledge on biodiversity. Two core BiCIKL partners (Pensoft and Plazi) are well-established and successful SMEs with a proven history in developing globally unique solutions for data liberation from literature and semantic publishing. Furthermore, the project will make use of the private sector knowledge amassed by two project partners (EBML-EBI and ELIXIR) as part of their established and vibrant industry engagement programmes. |
Sections "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "The BiCIKL contribution to fostering a culture of international cooperation", "Innovation is the key for BiCIKL to establishing a successful new community", "Expertise and complementarity", "Industrial and commercial involvement"; NA-03, JRA-01, JRA-05; Outcomes D6.1-6.4. |
|
|
Composition of the consortium |
Mobilise a comprehensive consortium of several key research infrastructures in a given field as well as other stakeholders.
|
BiCIKL starting community encompasses highly advanced RIs, including distributed ones (e.g. ELIXIR, DiSSCO, LIFEWATCH and GBIF), however operating in five separate and largely isolated domains of biodiversity knowledge. BiCIKL RIs are carefully selected to cover the entire research and data life cycle:
The current limitation of all these RIs is that the data linkages between them, and the provision of access to interlinked FAIR data, are still in their infancy. Non-interoperable and non-linked data do not allow a proper inter-disciplinary research on pressing Grand Challenges such as the loss of biodiversity and derived ecosystem services. BiCIKL will address these limitations by focusing not only on linkages between data classes, but also on means of curation and access to interlinked data, resulting in real-time research work. |
Sections "BiCIKL contributing to the ESFRI Roadmap", "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "Methodology", "Expertise and complementarity"., Fig. |
|
|
Stakeholders |
Main users: Researchers and research institutions. |
Biodiversity and genomics communities are among the largest ones in the life sciences. Many research areas dealing with organisms at different levels, e.g. agriculture/farming/aquaculture, biomedicine, nature conservation, environmental science etc., actually need and use data provided by the participating RIs, especially such on species names, sequences and literature data about these. Each of the data classes and their supporting RIs have strong communities of researchers who are actually the main users of their data holdings:
|
Sections "Objectives", "Improved access via BiCIKL as a basis for cross-cut research", "The BiCIKL community in a ten-years perspective", 1.4.3, 1.4.5; NA-01-03; JRA-01-05, TA, VA; Outcomes D4.3-4.4, D5.1, D6.3-6.4, D11.5. |
|
|
Stakeholders |
Other: Public authorities, technological partners. “Integrating Activities should, when relevant, contribute to fostering the potential for innovation, including social innovation, of RIs by reinforcing the partnership with industry, public administrations and/or other stakeholders, through e.g. transfer of knowledge and other dissemination activities, activities to promote the use of RIs by industrial researchers or policy-makers, involvement of industrial associations in consortia or in advisory bodies.” |
Data provided by BiCIKL will serve a wide range of users beyond science, including important policy making organisation at EU and national levels:
BiCIKL partnering SMEs will provide one of the key novel assets of the projects, namely FAIR data extracted from literature and next-generation publishing to:
|
Sections "Concept", "Methodology", "Industrial and commercial involvement"; NA-01-03, JRA-01; Outcomes D3.1-4.3, D6.1, 6.3. |
|
|
Country scope |
EU Member States, Associated Countries and other third countries when appropriate, in particular when they offer complementary or more advanced services than those available in Europe. |
The access provided by EU RIs will be open for interlinking and use for anyone, including leading international or national RIs:
Two partners are based in an Associated Country (Switzerland) |
Sections "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "BiCIKL as an entirely open science project, from start to end", "The BiCIKL contribution to fostering a culture of international cooperation", "Transformed access for a new community of users", 1.2.4, 1.4.2, 2.1.1; NA-03, Suppl. material |
BiCIKL contributing to the ESFRI Roadmap
In response to Priority 1 of v.d. Leuven Commission: A Green European Deal, and strictly following the guidelines and principles of the European Charter for Access to Research Infrastructures, BiCIKL will contribute to and participate in several ways in the European Strategy Forum on Research Infrastructures (ESFRI) Roadmap.
First of all, two RIs in the project consortium are classed as ESFRI ‘Landmark’ Research Infrastructures (LIFEWATCH, ELIXIR), which means that they represent major elements of competitiveness of the European Research Area. DiSSCo is itself classed as an ESFRI Project. BiCIKL is taking a scientifically oriented approach to bringing together European (including under the ESFRI Roadmap) and other infrastructures to provide new research pipelines to a variety of biodiversity scientists. In this context, the project is working in parallel with the activities taking place around relevant EU-funded activities such as the ‘cluster’ project EOSC-Life, coordinated by ELIXIR, and ENVRI-FAIR (ENVironmental Research Infrastructures building Fair services Accessible for society, Innovation and Research).
The existing clustering of Research Infrastructures is predominantly based on a domain approach (e.g. Environment for LIFEWATCH and DiSSCO, or Health and Food for ELIXIR) (Fig.
BiCIKL will advance the ESFRI Roadmap by clustering Research Infrastructures beyond domain barriers and improving their horizontal linkages in the projection of the Open Science concept (Fig.
Additionally, BiCIKL will support the further development of European infrastructures, which currently are not part of the ESFRI Roadmap, in their efforts to align with the requirements of the European Research Area and the premises and guidelines highlighted in its latest report dated December 2019, in which gender equality, geographical balance and optimal mobilization of scientific knowledge have a special focus.
For instance, BiCIKL shall ad-here to the principles of the European Charter for Open Access to Research Infrastructureacross all the participating infrastructures, overcoming both geographical boundaries by integrating a global approach as well as domain-approach by interconnecting RIs from different disciplines. BiCIKL will equally foster connectivity between research developments and (industrial) innovation by providing a sustainable, open, interconnected link to comprehensive data that will provide a much better understanding of the entire data cycle with no data leaks. The special effort dedicated by BiCIKL to training and capacity building will contribute to promote those endeavours on the long-term and foster strong engagement from different communities of users, including education. By encompassing all these 3 pillars, science and innovation together with education, BiCIKL strongly anchors on excellent research, market-driven opportunities and finally on provision of open data widely accessible and of knowledge transfer and circulation strongly supported, for the societal benefit at large.
The Work programme emphasises on the fostering of the long-term sustainability of research infrastructures and on expanding their role and impact in the innovation chain. All BiCIKL activities are actually in support to actions included in the 2016 Communication on the European Cloud Initiative, in particular to further integrate and consolidate e-infrastructure platforms, to connect the ESFRI infrastructures to the European Open Science Cloud, and to develop a European Data Infrastructures (EDI). To ensure better coordination and management of the innovation capacity of BiCIKL, Dr Donat Agosti (Plazi) will take the role of Innovation champion during the project duration.
By the end of the project, BiCIKL will have demonstrated how infrastructures that operate in adjacent but different ESFRI domains (e.g. Environment and Health and Food) can build strong technical and operational interfaces to support specific scientific needs that currently demand answers and also to contribute to open novel pathways for scientific developments.
BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process
Collections-based research and taxonomic science have been collaborative activities for centuries. Thousands of natural history collections worldwide share an established culture of cooperation to describe global biodiversity, exchanging specimens across borders and following international codes of nomenclature to maintain the vast taxonomic literature as a shared knowledge resource for use by all. This culture primed biodiversity researchers to become one of the earliest and most enthusiastic users of the Internet. For example TDWG (the Biodiversity Information Standards body) has been active since 1985.
As a result of these cultural foundations, thousands of databases and projects have been developed around the world to support the needs and interests of subcommunities of biodiversity researchers, organised along national, regional, taxonomic or thematic lines and variously managing different subsets of the potential knowledge space - species and other taxa, specimens, sequences, images, literature, distribution data, directories of expertise, etc. Many of these initiatives predated modern approaches to open data and open science. Although social expectations within this outward-looking community have favoured precisely the directions adopted for recent research infrastructure investments and more broadly for research cooperation (e.g. the Strategy for EU international cooperation in research and innovation), it has proven slow and difficult to reengineer this rich but fragmented landscape into a unified whole (
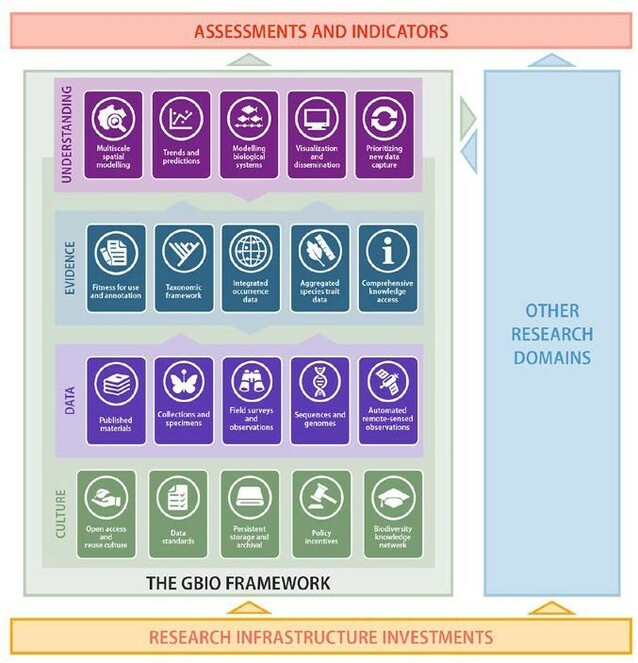
Large international reviews (
The second Global Biodiversity Informatics Conference (GBIC2) in 2018 reviewed the sociological and technical changes required to implement this vision (
BiCIKL is a response to the goals of the Alliance. It builds on the work of prior projects to address the fragmentation of European biodiversity and genomics databases and projects (BioCASE, EDIT, EU BON, OpenUp!, PESI, ViBRANT, pro-iBiosphere). These projects have advanced the maturity of the community of European natural sciences research-based organizations, working together in networks such as CETAF. They have therefore prepared a stable ground for enhancing the capacity of the community to collaborate and interop- erate at European level and also internationally, thus facilitating global user access.
The European and global infrastructures contributing to BiCIKL are committed to accelerating digital access to biodiversity knowledge, developing robust cross-linkages between different classes of data, and functioning as partners and exemplars for broader international standardisation and interoperability within the Alliance. This will ensure that developing infrastructures at the European scale, particularly DiSSCo, ELIXIR and LIFEWATCH, form world-class hubs in a truly interconnected landscape with other regional initiatives such as iDigBio, the Atlas of Living Australia, SANBI, CONABIO and ABCDNet. Developments within BiCIKL will also enhance and contribute to other European infrastructures that focus on the environment and ecosystems, including eLTER.
BiCIKL as an entirely open science project, from start to end
The concept of open science is aimed at transforming the way research is produced, accessed and utilised towards creating a primarily collaborative rather than a primarily competitive endeavour which works for the benefits of both scientific advancement and societal well-being, including developing economies in the Global South (
In line with this philosophy, most of the project’s outputs, including those produced at the early stages of the project’s life, will be published as a collection of articles (spanning grant proposal, methods, software tools, data, workshops reports, guidelines, and policy briefs) in the open science RIO journal, thus providing full transparency and reusability of these (see, for example, the open science collection of the EU BON project: Building the European Biodiversity Observation Network (EU BON) Project Outcomes). BiCIKL will go beyond providing open access and open data, towards establishing an open science research community and related services in the domain of biodiversity. The project aims to fully FAIR-ify biodiversity research by providing access and tools for linking and discovering data, publications and other intermediate results along the full line of the biodiversity research cycle: specimens → sequences → species → analytics → publications → biodiversity knowledge graph → re-use. This will contribute to making data and research produced verifiable and reproducible, which are cornerstones to reliable science. All activities in this important direction will be coordinated by an Open science champion, whose role and responsibility will be taken by the project coordinator, Prof. Lyubomir Penev (Pensoft).
In addition to fully opening up the research cycle, BiCIKL will further expand its measures for reusability and accessibility of results by providing project developed tools and software as open source, under appropriate licenses. The project will allow the community of users and stakeholders to freely use these tools, but also to further develop and enhance them.
By providing this, BiCIKL will conform to all fundamental principles of open sciences: open access, open data, open source, human- and machine-readability, interoperability, transparency and reproducibility of the full research cycle.
Concept and methodology
Concept
The BiCIKL conceptual framework is based on several most recent paradigmal changes in the way research is being performed, published and re-used:
- Open Data: Data on biodiversity should be openly and freely available to researchers and the society in general (see also the Bouchout Declaration).
- FAIR Data: Open data should be preserved and managed in ways that ensure Findability, Accessibility, Interoperability and Reusability (for FAIR data in biodiversity, see
Lannom et al. 2020 ).- To make data Findable means these to be assigned globally unique persistent identifiers, specified ithe metadata, described with rich metadata and registered/indexed/exposed in/to searchable resources.
- To make data Accessible means these to be retrievable through standard, free and open protocols, ideally at both the level of datasets and/or individual data records; metadata should stay accessible, even when data is no longer available.
- To make the data Interoperable means they should be identified, formatted, stored and described with appropriate metadata following formal, accessible, shared, and broadly applicable language for knowledge representation; interoperability requires also using of FAIR vocabularies and metadata to contain references to other metadata.
- To make the data Reusable means data should be available through clear and accessible data usage li-censes and provenance records, and compliant with domain-specific, community-accepted standards.
- Linked Data: Data representing different data classes, both within and beyond the domain of biodiversity, will be linked using community-agreed vocabularies and standards that enable participation in the Linked Open Data Cloud.
- Open Science: The concept of Open Science is the last stage of development of the “open triade” Open Access → Open Data → Open Science going far beyond providing free access to the published articles and data that underpin these, by assuming that all stages and outputs of the research cycle should be made open, transparent, reproducible and reusable.
Based on the above conceptual approaches, BiCIKL will ensure that its participating RIs will conform to:
- Providing free and open access to digital resources about biodiversity and associated access services.
- Pursuing use of licenses or waivers that grant or allow all users a free, irrevocable, worldwide, right to copy, use, distribute, transmit and display the work publicly as well as to build on the work and to make derivative works, subject to proper attribution consistent with community practices.
- Developing and applying policies that will ensure free and open access to biodiversity data.
- Improving tools and creating services for usage tracking of identifiers in links and citations to ensure that sources and suppliers of data are assigned credit for their contributions.
- Implementing indexing services and associated registers for content to allow discovery, access and use of open data.
- Promoting and implementing the use of persistent identifiers for digital and physical objects such as specimens, images, sequences, taxonomic names, treatments and literature with standard mechanisms to take users directly to content and data, both at the level of data sets and/or individual records.
- Establishing standards and relevant infrastructure for discovery and curation of bi- and multi-directional links between data across the data domains related to biodiversity.
- Refining the concept, priorities, technical requirements and application of a sustainable Open Biodiversity Knowledge Management System (OBKMS), that is attentive to scientific, sociological, legal and financial aspects.
To achieve the above conceptual goals, BiCIKL starting community has been assembled from well-established, European and global-scale research infrastructures that, although differing in origin, operational models, geographical locations, etc., all are globally significant in terms of uniqueness of services and data holdings. The coherence and enforced integration between the infrastructures will directly contribute to the restructuring of the European Research Area in the field of biodiversity into an ERA-based, overarching ecosystem of services operating across the entire biodiversity data life cycle.
The basic principle of forming the starting community is inclusivity, going far beyond the list of partners (beneficia- ries), participating in the project. The global ambitions of BiCIKL will result in a global approach in providing access across national and continental borders, hence the list of partnering RIs is complemented with other, critically important RIs from all spectra of the biodiversity research cycle. The participating RI are listed in Table
EU-based and global research infrastructures participating in the starting community.
|
Knowledge domain |
Research Infrastructures participating in BiCIKL |
Research Infrastructures linked to BiCIKL (see Suppl. material |
|
Literature |
BLR, Europe PMC, TreatmentBank, OpenBiodiv, ARPHA, SIBiLS, Zenodo |
BHL |
|
Specimens and samples |
GBIF, DiSSCo |
GGBN |
|
Genomic sequences |
ELIXIR, ENA, PlutoF |
iBOL, GGBN |
|
Taxon names and OTUs |
CoL, PlutoF |
iBOL |
|
Analytics |
SIBiLS, LW (e-Infra, BE_VREs) |
mBRAVE (iBOL) |
The level of integration between RIs will be based on the principle of minimum required metadata mapping and linking between RIs, which will ensure the access to the researchers’ user groups will be provided in two main modes:
- bi-directional linking and access between RIs (for example, specimens available through DiSSCo will be linked to sequences deposited in ENA), and
- multi-directional linking and access across data domains (for example, a specimen used and cited in a publication is linked to its digital object in DiSSCo, specimen occurrence record data in GBIF, sequences, extracted from it in ENA, and species name it belongs to in CoL).
With this, BiCIKL will ensure an unparalleled multiplication of data re-use across several biodiversity-linked domains, including data liberated from published narratives.
The BiCIKL community will directly benefit from conceptually novel approaches and technological innovations in managing and publishing biodiversity data, for example the Barcode Identification Numbers (BIN) of BOLD/iBOL (
In conclusion, the innovative character of BiCIKL can be seen at several levels and actually at each participating RI, however those than can be classed as globally unique, novel, cross-domain approaches are:
- linking to the actual collection specimens, biosamples, sequences or taxon names from the literature where these data elements have been cited;
- seamless conversion of published legacy narratives into structured data and linking these back to the original data sources, and
- cross-domain access to interlinked FAIR data.
Methodology
General approach
The BiCIKL programme of work includes several interlinked activities, ensuring improvement and provision of different levels of access to RIs within and across data domains, hence requiring different methodological approaches, carefully designed to form a coherent and coordinated set of components to deliver the BiCIKL objectives:
- Time-scale methodology: BiCIKL will start with identifying and describing the status quo of the level of FAIR-ness at each of the RIs. Most of the analyses performed within the Networking Activities activities will be delivered in the form of standards and guidelines not later than the end of the first project year, giving enough room for JRA development and access provision during the second and third year of the proposal.
- Bi-directional linking: BiCIKL will implement solutions to achieve bi-directional linking and metadata provision between RIs as standard practice, enabling each cited infrastructure to cite the citing infra- structure in return (examples: specimen<->sequence, publication<->specimen, taxon<->publication, tax- on<->specimen). For example, named entities in literature (specimens, accession numbers, taxon names) should be linked to their data objects in the respective infrastructures; the infrastructure, however, should link back the corresponding data object from the publications where it has been used and mentioned, e.g. via DOIs of the article, treatment or image. The generalised model for achieving this bi-directional linking between infrastructures is as follows (see Fig.
4 and the explanation in the text):- Identify elements (usually text strings) associated with one data object (Object A) that indicate a reference to another data object (Object B). This stage is associated with the repository holding Object A and is likely to be specific to the class of Object A. Examples include:
- Species Treatment (Object A) includes (in Materials Examined sections) CollectionCode + SpecimenID + ScientificName + Date + Locality, forming a reference to a particular museum specimen (Object B).
- Short bibliographic record (Object A) refers to Publication (Object B).
- Text string interpreted as scientific name (Object A) refers to TaxonConcept (Object B).
- Use these elements either directly to look up the associated data object (Object B). In optimal cases the supplied elements form a good (compound) key and can be used directly to locate the object. In other cases, a workflow will be required to find the likely referent. This stage is specific to the class of Object B and will be delivered via APIs that intelligently supperoth human-in-the-loop methods to assert links, which will be hardened as persistent links or bindings between the objects. These may be unidirectionally embedded within Object A or embedded in both objects or stored in a third place (Zenodo) that will serve as a join table or a linkage broker. Such an approach will be based on bespoke efforts within each infrastructure. All three approaches can support bi-drectional user navigations. Having a third party storage would ensure that we have a place to keep all links regardless of the abilities of the infrastructures to add them internally.
- Identify elements (usually text strings) associated with one data object (Object A) that indicate a reference to another data object (Object B). This stage is associated with the repository holding Object A and is likely to be specific to the class of Object A. Examples include:
- Disambiguation: algorithms will be developed that not only compare the metadata of the entities to be linked, but also includes the metadata of the entities themselves, for example by extending the matching data to include bibliographic information, biographies, geographic data of locations, dates of many events and nomenclature. Only by doing it, the needed confidence and volume of linking will be achieved. Furthermore, this process will be cyclic and will build on itself so whatever we will build will have to be rerun regularly. Such workflows are still in their infancy, one of the first examples of such approach is the Bionomia platform identifying and curating links between specimens and persons who have collected and/or identified them.
- Conversion to and use of Linked Open Data: the data liberated from literature and linked to external data resources of the participating RIs, as well as the taxonomic backbone used by CoL and GBIF will be converted using R scripts into RDF according to the OpenBiodiv-O ontology (
Senderov et al. 2018 ) and stored in the OpenBiodiv biodiversity knowledge graph (Fig.5 ). - Multi-directional linking and FAIR data delivery: all participating RIs will commit to a two-sided process of both ingestion or delivery of FAIR data, including such in machine-readable formats and via Web services, based on the minimum required standard metadata matching principle. This requirement is reflected in the composition and sequential ranking of tasks in each JRA WP (Fig.
6 ). This critically important and innovative method of access to data across domains will be provided through federated search methodology using AI tools and services through a central discovery, linking and link assertion mechanism, called FAIR Data Place (FDP). The FDP will be developed in a separate JRA-05 WP to ensure its integrative character across domains and will also include annotation/curation interface and a third party storage at Zenodo.
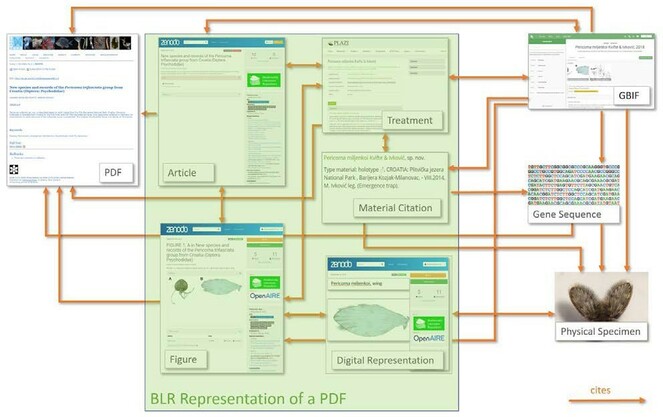
An example of the level of integration between the RIs that will be provided by BiCIKL is illustrated by the Biodiversity Literature Repository (BLR) workflow (Fig.
Once this process is finished, the file is stored in TreatmentBank and the entire article is deposited in BLR with rich metadata. In parallel, the figures and taxonomic treatments are also uploaded with rich metadata, and interlinked between each other and with the publication deposit. The obtained DOIs are then assigned back as attributes in the respective annotations in the stored file in BLR. This makes parts of the publications independent data objects that include links attributed with the metadata of the citations (e.g., a figure citation includes the DOI of the figure deposited in BLR; a bibliographic citation includes the entire bibliographic reference as attribute). Together with the assigned PIDs, this makes the data truly re-usable.
In a follow up step, GBIF is notified that a new publication is available and a new data set is to be imported as a Darwin Core Archive (DwC-A). GBIF gets the DwC-A and ingests the data which become available as a data set, taxonomic treatment and taxon occurrences. Each of these elements bears provenance data, which guarantees that the source PDF is always cited as well as are the FAIR data object deposited in BLR and, in fact, creates a multitude of links back to the source. At the moment that a publication is curated and changed, a new upload to GBIF is triggered. This set up allows users to upload and maintain the data sets at GBIF within minutes of processing in TreatmentBank. Once a day, a list of all newly described taxa (organisms sensu NCBI) are submitted to NCBI’s taxonomic backbone service. The proposed bi-directional linking in BiCIKL will assure that the citations do not just lead from the liberated data to the respective RI, but that they also cite the data in BLR.
Project structure
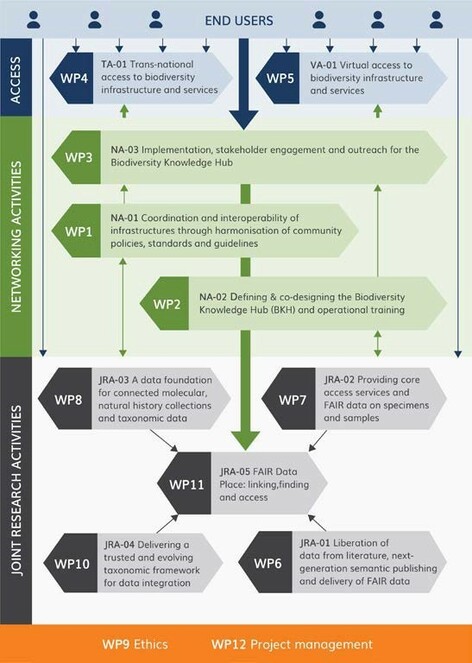
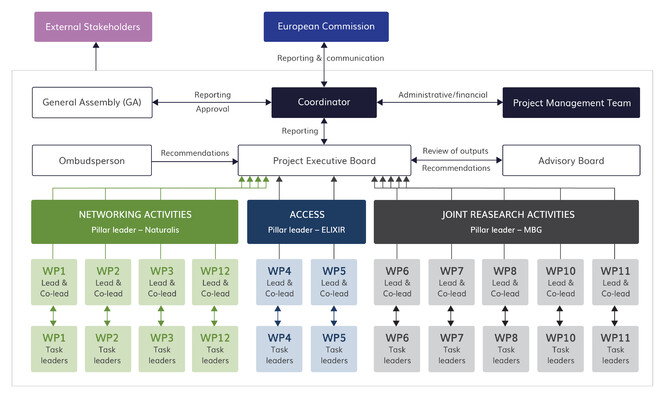
To help navigate this complexity, activities have been structured into three pillars, representing the three RI activities: NA, JRA and TA/VA. Each will have a pillar leader whose role is to oversee the work and interlink activities with the work of other pillars (see Fig.
Networking Pillar (NA)
The NA pillar will coordinate and optimise the integration and harmonised access between RIs and their respective data classes, as well as engage stakeholders, improve capacity through training, communicate and disseminate the project access tools and services. The NA pillar will be led by Partner 2 Naturalis and will consist of three work packages:
NA-01 Coordination and interoperability of infrastructures through harmonisation of community policies, standards and guidelines comprises networking actions addressing the policies, best practices and standards that our scientific community needs to be aware of, adopt and implement at institutional level. This will constitute a thorough, harmonised knowledge base on which further collaborative developments can be built and will be complemented by development of new or underspecified standards to make them suitable for the needs of data exchange and interoperability between infrastructures. NA-01 will review user requirements from the community in terms of the competent questions they want to address with the data and ensure that the data access methods, such as websites, downloads and APIs, conform to user requirements and are optimum for the volume of requests they will receive. At the end, NA-01 will produce aligned data sharing policies and access procedures to ensure easy use and transparency to the end user, whilst at the same time monitoring and reporting on use.
NA-02 Defining and co-designing the Biodiversity Knowledge Hub (BKH) and operational training. The starting community needs first to be identified based on its own novel nature, then reinforced internally and finally supported in its functionality. Separate endeavors currently being undertaken in isolation need to be aligned and merged among different RIs and mechanisms need to be defined and implemented to ensure its sustainability and cohesiveness. All together will be reflected by the identification of the elements of the BKH that will be followed by the specifications for its implementation of the BKH further developed in NA-03. Once the working model is defined, a comprehensive training programme will ensure capacity building addressing the needs of the community. It will provide the appropriate tools and best practices to allow access to services at all stages of the data cycle, from generation to analysis and final use.
NA-03 Implementation, stakehold- er engagement and outreach for the
Biodiversity Knowledge Hub. To build the new Community, it will be key to establish and maintain an active dialogue between the participating RIs with other stakeholders (researchers, libraries, publishers, digitization technology providers, digital repositories, research institutions and public authorities), through a modern, multi-functional, communication and publishing platform, serving also as a knowledge broker for the end users. This role will be first performed by the project website firstly integrated into the BKH and later hosted by GBIF after the project lifetime. The platform will be operating on several levels acting as an information cluster (internal communication and feedback system, publishing platform for community-related documents, including reports, standards, guidelines, policy briefs, social media profiles) to ensure effective integration, prioritization, cost effectiveness and sustainability of the community’s communication interface, networking activities and operations during and especially beyond the project. The engagement flows with both providers of data and users of the derived services will be set up through a series of expert workshops and sessions, bringing together the experiences of key parties.
Access Pillar (Trans-national and Virtual Access, TA & VA)
The new BiCIKL community will provide two main forms of advanced access, Trans-national, to named users (TA) and Virtual, to unidentified users (VA), each of which has a specific role in achieving the project objectives. The pillar will be led by Partner 5b ELIXIR Hub.
TA-01Trans-national access to biodiversity infrastructure and services will be focusing on services that support efficient integration of diverse data sets across data domains to meet the needs of user groups of named scientists, who will apply for provision of specific services via open call projects that will require special efforts at the RI side. The open call criteria will be formulated to support innovative methods of access, for example to linked data across domains, for example provided by at least two of the participating RIs, operating in different data domains. TA will encourage provision of services to user groups from country(-ies) different from the host country of the RI installation which will be provided through remote trans-national (no visits needed) access in order to encourage use of virtual research environments.
VA-01 Virtual Access to biodiversity infrastructure and services is suited for the basic Open Science concept of the BiCIKL community, namely harmonised access to biodiversity data that are Open, FAIR and Linked, by not requiring identification of users (except for registration/login purposes where needed). Thus, VA will ensure a large-scale, barrier-free internationalisation of the BiCIKL products and services, hence contributing to the global leadership Europe has had historically in biodiversity research through the world’s oldest and richest natural history collections and derived data. Provision of greater and more flexible VA will exploit the existing services and especially those improved or newly developed in JRA, with EU financial support only being used to cover the technological and sci- entific support for the VA access activities needed by researchers.
Joint Research Activities Pillar (JRA)
The new starting BiCIKL community can only be built and made sustainable if the data domains and their operating RIs develop an efficient system for
- sharing,
- linking and
- providing FAIR data to each other and to re- searchers.
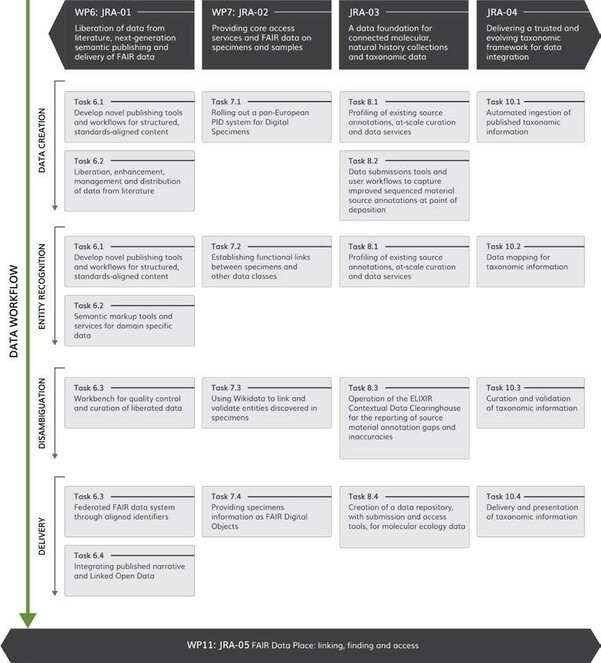
To ensure this critically important condition for the success of the project, the JRA activities are designed to improve each RI’s interface and background technologies so that to allow both ingestion and provision of linked data, while following a generally unified concept for activities within each JRA WP along the logical construction: Data creation -> Entity recognition -> Disambiguation -> Delivery (Fig.
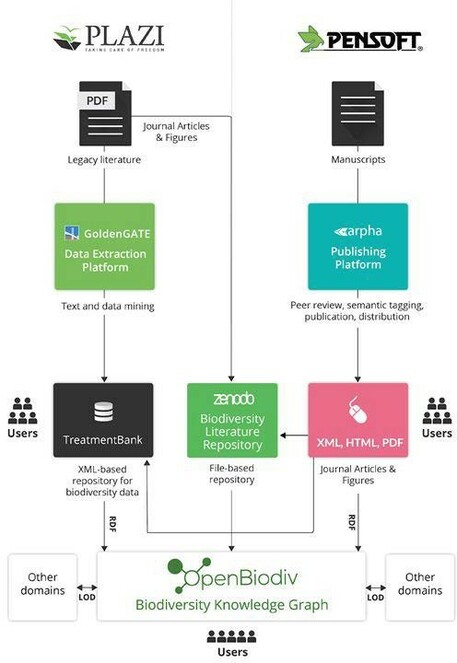
JRA-01 Liberation of data from literature, next-generation semantic publishing and delivery of FAIR data is at the core of the new BiCIKL community and a definite and unparalleled novel contribution to the biodiversity data landscape. The BiCIKL community will create and exchange free flowing workflows of data extraction, enrichment and access, including next-generation publishing tools that ensure provision in both human-readable and machine-retrievable, semantically enhanced, structured data extracted from the lit- erature (Fig.
The data publication, independently of its format, will be ensured to have adequate representation of its semantics and to be citable with persistent identifiers either assigned during the publishing process, or retrospectively, during the text and data mining process. That is to say, we will represent what a data element means and how it should be interpreted by machines. From the viewpoint of the Open Science principles, JRA-01 will provide a holistic approach and routine services for inclusion of FAIRified literature data, extracted from various sources (across history, journals and publishers) back into the research life cycle. Moreover, it will also provide the critically important link of interoperable Linked Open Data across biodiversity and with other domains via the RDF-based, OpenBiodiv Biodiversity Knowledge Graph.
JRA-02 Providing core access services and FAIR data on specimens and samples. In spite of the critical importance of the natural history collections as a fundament of the biodiversity knowledge, less than 10% of the museums’ holdings are currently digitized (
JRA-03 A data foundation for connected molecular, natural history collections and taxonomic data. In the molecular biology databases, for example the European Nucleotide Archive (ENA), the ELIXIR Core Data Resource in which primary nucleic acid sequence data from specimens and environmental samples are maintained, the relevant annotations relate to the natural history collection, biobank or culture collection source of the organism that has been sequenced and the accession codes for these sources. While such annotations exist for many sequence records, they are neither complete (many sequences are not linked to their sources), unambiguous (many annotations do not lead to a single endpoint relating to the source), nor necessarily accurate (many incorrect annotations exist). JRA-03 will establish a data foundation for connecting sequences, collections, taxonomy and literature through at-scale curation and organisation of molecular biology data and services. This key objective will be achieved through building user tools and workflows that drive accurate reporting of source annotations into molecular biology databases at time of deposition and facilitate data update cycles in case of inaccuracies. The WP will also enable structured scholarly publication of molecular ecology data insufficiently served by current infrastructures.
JRA-04 Delivering a trusted and evolving taxonomic framework for data integration is based on the understanding of the central position of taxonomy in managing biodiversity information and how this affects almost all associated user interactions, either as a primary filter on data matching a request or as fundamental information for interpreting results. JRA-04 focuses on FAIR improvements to all stages in the process to harvest, aggregate and curate taxonomic information from publications and genomic research. This includes accelerating standardised access to streams of new or digitised species treatments and MOTU (Molecular Operational Taxonomic Unit) classifications, mapping of all of these into a consolidated Catalogue of Life (initially dependent on many decisions from automated processes), offering initially automated mappings between CoL and other significant species checklists, enabling expert community curation of all these derived products, and encapsulating these in services and visualisations that support the infrastructures of all BiCIKL partners and other users.
JRA-05 FAIR Data Place: linking, finding and access builds on the critical importance of convergence on standards and processes and data linking when digital records are to bridge across different RIs in different data classes to achieve their full scientific potential (Fig.
JRA-05 will leverage different approaches, including AI-based tools, and resources as delivered by JRA-01-04 and the participating RIs, to facilitate the seamless interoperability between data domains to allow much more rapid and repeatable access to data. JRA-05 will bring BiCIKL partnering and collaborating RIs’ assets together using novel technologies for discovery, validation, and preservation of bi- and multi-directional links within a central FAIR Data Place (FDP) service. Building on existing standards for management and use of linked data, JRA-05 will also draw on other fields that use biodiversity and -omics data, for example invasion biology, conservation ecology and climate research.
Integration between pillars
Integrating JRA with TA/VA: BiCIKL builds upon new levels of access that centre around two main points:
- FAIR data exchange and
- access to linked data across data classes.
The entire JRA is dedicated to laying the ground for these novel methods of data re-use. For example, one of the key requirements for approval of Open Call projects is to use data that are linked between at least two or three data domains, and JRA is meant to provide exactly that feature. Besides, JRA-01 will offer a unique new service, further tested and implemented through TA, that is a large-scale text and data mining from both historical and recent publications dedicated to a particular taxon or research topic. Digital access to data liberated from literature and linked to the RI data sources will enable poten- tial users to make better informed choices and research plans targeted, and generate new hypotheses and knowledge deriving from the big data pool assembled together from different resources. TA/VA users will also be able to use the improved technologies for multi-directional data discovery and linking through the FDP developed in the JRA-05. Another long-standing problem with proper disambiguation, validation and curation of interlinked and annotated data will be resolved at each RI and also centrally via the curation workbench developed in JRA-01 and JRA-05.
TA/VA users will be asked for their feedback on the development of the JRA outputs, for example on the functionality of the FDP service and the usability of the complex datasets derived from it for research purposes. This feedback will be used to refine the JRA outputs to better meet user demands.
Integrating NA with TA/VA: NA-01 work on digital standards and processes will increase the interoperability of collection data and services, speed up services, discovery and delivery of data to users. The harmonisation of policies in NA-01 will raise the standards of linked data preservation and accessibility, thus ensuring maximum accessibility by both current and future TA/VA users. In addition, NA-01 will identify areas of unrealised weakness in the cross-domain linkage and access to data through the continued and expanded use of TA assessment tools and via user feedback from the TA/VA users. NA-02 will support TA/VA through the identification of the required elements to cover the entire data cycle and the interlinked derived services and will tackle both TA and VA needs by supporting the community in acquiring digital (data) skills and competencies that enable users to navigate complex datasets effectively, as well as optimise curation and validation of linked data. NA-03 will support the development and operation of the Biodiversity Knowledge Hub (BKH) by leveraging the various needs and requests from the community identified in NA-02 into consistent and coherent specifications for knowledge brokering through the BKH one-stop entry point. TA/VA users will be made aware of all new developments and guidelines developed and refined in both NA-01 and NA-02 via an efficient communication and dissemination strategy and implementation plan provided by NA-03.
Integrating NA with JRA: These two pillars will be working closely together from the very start to the end of the project, which will be reinforced by a management decision for each of NA’s and JRA’s pillar leaders to also lead one work package in the other pillar. Several JRA activities are expected to start after the 6th month of the project after provision of feedback from the surveys on user requirements and interoperability hackathons performed by NA-01 and partly by NA-03. The feedback on newly developed or optimised JRA services will continue through the project, especially through the training and networking activities in NA-02 and NA-03 respectively, but also in a close collaboration with the TA and VA WPs. The JRA services, especially those that provide new features or federated data access will be described in guidelines developed in NA-01 and communicated to the users in NA-03, especially through the BKH that will be identified, described and analysed in NA-02 and further implemented in NA-03.
Ambition
The fragmentation of the biodiversity informatics and how BiCIKL will improve it
Knowledge of European (and global) biodiversity is currently spread across hundreds of databases, across different data domains (see
- at each participating RI and
- developing interoperable linking systems between them,
using agreed standards, tested and implemented in TA and VA.
A good illustration of the “fragmentation” problem is the ever-increasing gap between
- the innumerable and ever increasing number of sequences (including whole genomes) and
- historical (e.g. collection) data connected to the Linnean species, leading to the “dark taxa” problem (
Page 2016 ).
There is an entire world of RIs that link sequences to taxon names and specimens (INSDC, BOLD, UNITE, SILVA and others), however linking to the actual collection specimens or biosample, let alone linking to the literature where either the specimen or its sequences have been cited, requires focused, coordinated and extensive development. BiCIKL will provide improved access through API-based service to identification and disambiguation of links between sequences and collection specimens, through usage tracking of sequence accession numbers and specimen IDs at both, the level of data infrastructure and in the published literature. In the latter case, BiCIKL will provide access to taxon names and their treatments extracted from literature and linked to the respective specimens and sequences these are based upon.
Another fundamental problem is the inefficient use of and access to the invaluable data imprisoned in legacy literature published on paper and PDF, a process that is often called “PDF impediment”. The huge domain of legacy literature seems to have fallen outside current technological developments, despite large-scale digitization efforts, such as BHL. Whilst PDFs, or some elements in them, can be identified and accessed with software tools, the community is still far behind converting the literature into structured data, hence making it really usable, especially to a new generation of digitally literate scientists. The newly published literature, if only in PDF, also contributes to the reduced usability and inaccessibility, instead of resolving it, because data inside PDF text can neither be easily accessed nor indexed. Still, the domain of biodiversity has one of most advanced data and text extraction and publishing infrastructures in the world (Fig.
Despite the concerted effort of the entire biodiversity community that has resulted in the Catalogue of Life (CoL), we still lack a comprehensive, up-to-date, community-agreed list of known species on Earth. There is an entire host of problems connected to the stability of names and their disambiguous linking to taxon concepts, specimens and sequences. BiCIKL is targeting the problem through building up an automated alerting system to inform CoL, GBIF and other aggregators about newly published taxon treatments, including new taxa. CoL will create a service to ingest new taxon names, and expose the nomenclatural changes to community curation.
Due to the heterogeneity of research on biodiversity, the network of experts is divided into small groups studying a particular taxon, often working “independently” from each other. Another cause of segregation is the division between the biodiversity and genomics communities, which is reflected by the way data are handled, accessed and used by these two communities. BiCIKL’s key goal is to bridge these fragmented communities by putting in place highly automated ways to link sequences with specimens, literature and names, as well as by encouraging and supporting clear cross-disciplinary aspects of the TA open call applications (an example for such an approach is Use case #5 in Suppl. material
The BiCIKL contribution to fostering a culture of international cooperation
BiCIKL will close a gap between data domains and improve interlinked access to their holdings, thus contributing to a global effort towards establishing open science practices that provide FAIR biodiversity-related data. Work packages are led by the major global standards and data sharing organisations (GBIF, ELIXIR, EMBL-EBI, CoL), coupled with the networked power of the European natural science institutions (CETAF) and technological partners (SIB, UTARTU), including such from the private sector (Plazi, Pensoft) that altogether will help sustain a culture of cooperation between project participants and the international community. Working closely with the leaders of comparable international efforts (e.g. iDigBio, BHL, GGBN, iBOL), and building on efforts to construct the next iteration of the Alliance for Biodiversity Knowledge (
Implementing such a mechanism will allow BiCIKL and other regional and national initiatives to benefit more effectively from distributed international investments. As a truly international undertaking, this is essential to achieving our community’s long-term ambitions.
Improved access via BiCIKL as a basis for cross-cut research
BiCIKL will harmonise digital access to several world-class data infrastructures and will work towards providing linked data from various domains, with the clear goal of setting the ground for next-generation research. Through new methods for access to data and services, developed in JRA and implemented in TA/VA, including aggregation, linkage and curation of data, critical new insights will enable scientists to address some of the world’s greatest challenges. Our community’s ambition is to provide the data and tools to support consistent and comprehensive global discovery and use of information from all sources about the biodiversity of any defined area, over time, and covering all taxonomic groups. Within BiCIKL, the development of the FAIR Data Place (FDP) (under JRA-05) will prove a unified federated search and access, combined with efforts to increase standards compliance, especially by implementation of linked open data capabilities, and artificial intelligence. The linking mechanism for stable identifiers across the data RIs, including literature, is a major step towards achieving this ambition. This will create a mechanism by which users can discover data they would not otherwise find. In the long-term, this will allow the community to work towards all the information around specimens, species, sequences, taxonomic names, and literature being managed and curated as an inter-connected digital knowledge base. This work will also have the effect of creating a critical mass of highly skilled researchers focused on major scientific and related societal questions, which can be illustrated with the following examples (see also Suppl. material
- Biodiversity and ecosystems are currently under unprecedented pressure and transformation driven by extreme environmental changes. The cross-linking services and access to data from various resources, including historical data from collections and literature, will provide the basic knowledge to model, predict and recommend mitigation measures for the effect of global changes. There is also a rapidly-growing interest in innovative infrastructure services for early warning indicators (including fast-tracking of biological invasions) for which BiCIKL has the full potential to aid scenarios and tools for decision-support and environmental management, based on the innovative use of cross-domain data resources.
- Biodiversity and ecosystem services underpin all our life and the proper functioning of the planet as a whole, besides provision of food and many other materials. Integrated European RIs will provide new ways to study the properties of biological and genetic materials and resources. Related to this, cooperation organisations, such as the European Life-sciences Infrastructure for Biological Information (ELIXIR), which is a key BiCIKL partner, will provide scientists innovative and sustainable ways to develop and design such materials.
- Most economic sectors closely linked to nature, many of which are considered to be some of the fastest growing sectors of economy (e.g., recreation and tourism, land use, environmental assessment and health, energy production, mining, agriculture/forestry, bio-economy and many more) are dependent on biodiversity and natural resources in one way or another (
Kelling et al. 2009 ,Purves et al. 2013 ). Sustainable management of our natural environment is not possible without monitoring and prognostics based on reliable and readily-available data using cost-efficient, “real-time” methods, to which BiCIKL will directly contribute via sophisticated new tools for delivery of data on sequences, species, specimens and literature to support novel metabarcoding and metagenomics techniques. - Natural disasters, such as pandemics, can be directly connected to biodiversity, as it became apparent with the recent Ebola, Zica and especially COVID-19 outbreaks. Vectors or hosts of the pathogens are often wild animals, which raises many questions on why formerly unknown disease agents managed to spread so quickly among people. It is more than obvious that combating these huge challenges is impossible with medical or epidemiological means only; it should definitely mobilise both recent and historical data on biology and interactions of vector species with their biotic and abiotic environment.
Innovation is the key for BiCIKL to establishing a successful new community
BiCIKL will undertake Joint Research Activities necessary to deliver innovative solutions that have never been possible before for:
- two distinct classes of service provided by each participating infrastructure (inbound and outbound linking),
- interlinked access to standardized data, complemented with
- support for each of the above services through helpdesks, dashboards and training for biodiversity and genomics scientists.
These novel approaches are illustrated through five use cases in Suppl. material
Fundamental to achieving the ambition of BiCIKL are the means to deliver vast quantities of digital information from European and global RIs in different, previously fragmented data domains. Using advanced machine learning processes employing computer vision and artificial intelligence methods, the FAIR Data Place (FDP) built in JRA-05 is intended to enhance the discoverability of data located in various places and especially to provide, store and curate links between them. This platform will also provide human-in-the-loop functionality allowing experts and members of the public to improve the automated processes and enhance records. This cutting-edge innovation, the basis of which was established through work in several previously funded EU projects, will have a transformative effect on the biodiversity and genomics communities, putting ahead the community ambition of having an integrated knowledge base for supporting cross-cutting research. This innovation supports the BiCIKL ambition of centralising access requests and will be applied to services of the TA and VA programmes of BiCIKL.
The radical transformation of big data usability will also require radical changes in the way research results and data are published towards machine-readable data and narrative-integrated XML-based formats. BiCIKL will tackle this issue by improving the publishing workflow that allows import of data from RIs directly into manuscripts (JRA-01). These data are kept in XML during the entire publishing cycle and delivered back to data aggregators and, hence to the common FAIR data pool.
Another key innovation of BiCIKL will be the development of mass digitisation processes and highly automated TDM workflows for data liberation from published narratives accumulated during more than 300 years of biodiversity exploration (JRA-01). Supported by technical innovations in areas of digitisation, data/media processing and artificial intelligence techniques for text, tables and images, and working with the SME partners, BiCIKL will promote industry solutions in these areas, responding to new requirements with new products and services. The liberated data will be made accessible in a sustainable way through our partner OpenAIRE Zenodo public repository based at CERN. The process will be advanced through technical interoperability and data exchange standards implemented between publishers and data aggregators. The unique result of all this effort will be the delivery of extracted and FAIRified literature data back into the data life cycle, where these data will be linked and re-used together with data from both other literature resources and raw data from the RIs.
Going further, data liberated from literature (JRA-01) and taxonomic trees improved and accessed via GBIF and CoL (JRA-04), will be converted into RDF (JRA-01, task 6.4) ultimately generating an immense LOD OpenBiodiv biodiversity knowledge graph that can be linked with other LOD resources or corpora of knowledge (for example PMC Europe or Wikidata) that will put Europe at the forefront of supplying authoritative information about the natural world. The knowledge graph services provided in JRA-01 (OpenBiodiv) and JRA-05 (FAIR Data Place) and supported within BiCIKL through standardisation activities (NA-01), will accelerate the transformation of e-infrastructure services in Europe towards the European Open Science Cloud (EOSC). This work will promote (partial and integrative) federation of European biodiversity, environmental and molecular biology RIs by providing the reference data required to operate their services and stimulate innovation drawing on the use of big data technologies.
The BiCIKL community in a ten-years perspective
Looking at the European policy priorities for the next few years, it becomes apparent that the new European Green Deal and the European Open Science Cloud (EOSC) have become focal points. Europe is calling on its scientific community to step up and deliver robust scientific tools and data, upon which decision making can rely. Research infrastructures find themselves at the core of this societal need, aiming at providing the infrastructure capacity needed. Access to individual resources and tools is, however, not adequate for our scientific communities to deliver at the scale and quality needed. A new integrated approach is required that enables researchers to seamlessly navigate the entire landscape of tools, services and data sources and that gradually lowers the barriers to use across infrastructures. Examples of such approach are use cases #1 and #2, dealing with rapid response mechanisms for data gathering on biological invasions and COVID-19 pandemic (Suppl. material
The diversity of data types biodiversity science is relying upon (i.e. from species occurrence records to complex genomic studies, taxonomic interpretations or biochemical analyses) highlights even further this need for a more integrative approach to existing (data) infrastructures. BiCIKL focuses on this exact need by developing technical and semantic bridges between infrastructures involved in the biodiversity data lifecycle. As this new starting community of infrastructures grows, it will further improve all aspects of the operational and technical interfaces among the key actors. It will do so, benefiting from global contexts, such as the Alliance for Biodiversity Knowledge, as well as the work carried out within the Research Data Alliance (RDA) towards the development of a disciplinary interoperability framework. Furthermore, BiCIKL will work closely with the relevant EC-funded cluster projects (such as ENVRI-FAIR) and benefit from the investments there towards a set of commons (resources and tools). The investments in EOSC will also further strengthen the ability of biodiversity infrastructures to interoperate, as they gradually rely on common e-infrastructures (e.g. for Authentication and Authorisation).
BiCIKL will have the challenging goal to pave the way and demonstrate the feasibility of interlinking information and services across infrastructures but also support users in accessing this new integrated landscape. At the end of the project we anticipate having in place a clear path towards which future investments can focus on the integration of the biodiversity knowledge space.
Gender Balance and geographical distribution underpins BiCIKL
BiCIKL considers gender equality an instrumental axis of action and a target to achieve and maintain along the project timeline and across all project activities. To that end, BiCIKL addresses it from two perspectives. Firstly, gender balance in the managing and advisory teams of the project will be sought by the inclusion of corresponding recommendations by the Project Executive Board in the rules of participation in the decisive teams of the project, within the Operational Procedures (WP12). Secondly, the project will appoint an Equality and diversity champion (Dr. Ana Casino, CETAF) who will be coordinating all aspects of gender and other equality issues during the project duration. Within the BiCIKL team, three of the 11 WP leaders are female (WP1, WP3 and WP12), two of the 14 organisations have female PIs, and around 30% of the project participants are female.
Furthermore, BiCIKL will not only look into the need to invite or select team members on a gender-balanced basis but will contribute to promote and develop gender equality in the training programme (under NA-02) with a special emphasis on targeting and thus enable women to better access and become integrated into the new community, from different perspectives: throughout the variety of actors involved, by supporting different positions also at scientific/managerial high level, by promoting the participation of women in all stages of the data life cycle activities and at any possible level of responsibility, by ensuring multidisciplinary interaction and connectivity among different RIs to foster women engagement in BiCIKL. Additionally, gender and other important elements of equality (i.e. on race or religion) will be considered and encouraged during the TA project calls envisaged in WP4.
The entire BiCIKL project will be implemented to ensure equality, but also to create an empowering environment for supporting diversity, by encouraging the active participation of researchers from different regions. Moreover, a geographic balance within the new community will be sought from its beginning, seeking active involvement across all RI partners. In that respect, the participating RIs will continue tackling this issue themselves and carrying out specific actions to reach out to new members and in filling geographic gaps for new partnerships in under-represented areas such as south-eastern European countries. In this respect, the participation of global RIs and their associated networks, such as GBIF, iBOL, CoL, BLR, TreatmentBank BHL, ARPHA-XML and others, will bring cultural and geographical diversity to the project by ensuring a barrier-free access and participation for researchers’ groups from anywhere in the world, with special attention paid to the Global South.
Impact
Expected impacts
Transformed access for a new community of users
BiCIKL has the full potential to transform the entire landscape of biodiversity-related research, even because the building blocks of the starting community consist of RIs which provide novel ways of access to interlinked FAIR data in ALL critically important biodiversity data classes: collections, sequences, taxon names and literature. Moreover, BiCIKL will link the RIs involved along the data targeted uses as to build up a seamless path of connected data, from specimen data to publications. BiCIKL will broaden the user base for biodiversity data beyond taxonomy or genomics themselves, it will rather bridge these two and other neighbouring communities, both geographically and across disciplines, by focusing and fully supporting unlimited VA alongside the TA activities (see Table
BiCIKL contribution to the impact expected by the INFRAIA-02-2020 Work Programme.
|
N |
Expected impact |
Contribution of BiCIKL |
Evidence of impact |
Addressed in the proposal |
|
1 |
Researchers have wider, simplified, and more efficient access to the best research infrastructures they require to conduct their research, irrespective of location. They benefit from an increased focus on user needs. |
BiCIKL will provide a principally new level of improved access to each of the participating RIs and through the Biodiversity Knowledge Hub to data and services across formerly fragmented data domains and virtual research environments, including literature data linked to specimens, species and sequences. Concretely, this innovation will save time to end-users as they will be able to seamlessly access, through the hub, a larger pool of enriched data and knowledge, thereby by passing the traditional barrier of having to search multiple databases and then link up different data types, or to manually extract data from literature. Online access to data and knowledge will enable more researchers to benefit, including those in developing countries, especially as collection specimens often come from these areas of the globe. |
Usage stats of the BKH and separate RIs by geographic location and subject areas of study. Usage stats of geographical and subject area distribution of the open call project applications. Number of inputs to the user requirement survey and analysis. |
Sections "The fragmentation of the biodiversity informatics and how BiCIKL will improve it", "Gender Balance and geographical distribution underpins BiCIKL", "Transformed access for a new community of users","Expertise and complementarity"; JRA-01-05, NA-03, VA, TA; Outcomes D1.1, D1.3, D3.4, D4.2-4.3, D5.2-5.4, D11.5; Use cases in Suppl. material |
|
2 |
New or more advanced research infrastructure services, enabling leading-edge or multidisciplinary research, are made available to a wider users community. |
The strong emphasis of BiCIKL on JRA’s improved, harmonised and newly developed services, will support researchers in their needs to ask complex, cross-disciplinary, data-driven and frontier research questions, for example:
|
Number of user requests for cross-domain data through the FAIR Data Place and the OpenBiodiv SPARQL endpoint. Number of open call cross-disciplinary applications. |
Sections "BiCIKL as an entirely open science project, from start to end", "Improved access via BiCIKL as a basis for cross-cut research"; NA-03, JRA-05, TA, VA, Suppl. material Outcomes D4.2, D4.3-4.4, D5.1, D6.4, D11.1, D11.4, D11.5.; Use cases #1 and #2 in Suppl. material |
|
3 |
Operators of related infrastructures develop synergies and complementary capabilities, leading to improved and harmonised services. Economies of scale and improved use of resources across Europe are also realised due to less duplication of services, common development and the optimisation of operations. |
Synergy, complementarity and inclusivity are at the core of BiCIKL open science rationale and philosophy. The integrative character of the project is guaranteed by a methodological framework focusing on bi- and multi-directional linking between RIs, in addition to its central focal point - a harmonised and integrated access to interlinked and re-usable data across domains. In other words, the project will be creating added-value information not previously available to users of the existing and various infrastructures. Beyond saving time to the users (who need neither to re-create this information nor to acquire the skills to do so), there are clear cost-savings to be realised by the infrastructures themselves through better integration at scale, less effort duplication, and maximised output for the same initial investment. More broadly, it is well-acknowledged that open science leads to significant cost-savings, with a recent report1 finding that the annual cost of not having FAIR research data costs the European economy at least €10.2bn every year. |
Documented synergies and complementary capabilities. Estimates of cost-savings by fast and efficient access to data. Evidence for multiple data re-use for different research purposes. Cost-savings from using the automated literature data liberation workflow. Cost efficiency estimate of the integration of the
|
Sections "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "BiCIKL as an entirely open science project, from start to end", "Transformed access for a new community of users"; NA-01, NA-03, JRA-01-05, Suppl. material Outcomes D1.2-1.3, D.6.1-6.3. |
|
4 |
When applicable, innovation is fostered through a reinforced partnership of research infrastructures with industry. |
A unique, innovative asset of the new community is provided by two SMEs ensuring an unprecedented access to data extracted and FAIRified from literature, through one of world’s most advanced infrastructures for structured and semantic publishing, data liberation, management and re-use. This innovation may be picked up by other disciplines (e.g. Social Sciences and Humanities) where significant information is imprisoned in literature. |
Quantity of liberated data; processed pages, treatments, figures and tables; number of user requests to BLR & Treatmentbank. Number of journals using semantic publishing workflows. |
Sections "Methodology", "Innovation is the key for BiCIKL to establishing a successful new community", "Novel virtual access services and support structures", "Industrial and commercial involvement"; JRA-01, TA, VA; Outcomes D4.2-4.3, D5.2-5.4, D6.1-6.4.; Use cases in Suppl. material |
|
5 |
A new generation of researchers is educated as to be ready to optimally exploit all the essential tools for their research. |
The innovative modes of access will focus on the forthcoming generation of researchers through training and networking activities, building upon the already widely established networks and capacity building experience of CETAF, DiSSCo, ELIXIR, and LIFEWATCH. This will contribute to building Europe’s human capital for the digital age. |
Number of training courses organised. Number of trainees. Number of downloads of guidelines and instruction pages from BKH. |
Sections "Novel virtual access services and support structures", "Dissemination and exploitation of results", "Communication activities"; NA-02, NA-03; Outcomes D1.3, D2.2. |
|
6 |
Closer interactions between a larger number of researchers around a number of RIs facilitate cross-disciplinary fertilisations and sharing of knowledge & technologies across fields, between academia & non-academic stakeholders, including industry. |
Relationship capital, i.e. the benefits of working together in terms of knowledge-exchange, is at the heart of this expected impact, and it will be an outcome of this project. In BiCIKL, the most valuable result towards a more integrated ERA would be to bridge the increasing gap between the organismic and molecular biology communities. A close collaboration with industry will be showcased by technological companies (Pensoft & Plazi) whose unrivalled tools, workflows and services for publication and handling of literature data will serve researchers’ needs for better and more efficient science. A good example of that is the COVID-19 Task Force recently organised by several BiCIKL partners (see: https://cetaf.org/news/joint-cetaf-dissco-covid-19-task-force-call-contributions and https://blog.pensoft.net/2020/04/07/plazi-and-pensoft-join-forces-to-let-biodiversity-knowledge-of-coronaviruses). Potential industry partners of BiCIKL will be actually all publishers dealing with biodiversity data and information which can use the publishing platforms developed by Pensoft and the TDM tools and workflows developed by Plazi. The BLR at Zenodo can be used by any journal or publisher to deposit their articles and data. The tools and workflows developed in biodiversity genomics will be part of the EMBL-EBI Industry Programme. BiCIKL will use also the DiSSCo user stories and programs to work with the private sector and, more generally, the overall framework of EOSC expanding to the private and public sector activities. |
Number of publications using interlinked data. Number of cross-disciplinary open call proposals. Number of proposals involving services provided by the SME partners. Number of non-academia (private or public) users of the BiCIKL services. |
Sections "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "Methodology", "The BiCIKL contribution to fostering a culture of international cooperation", "Improved access via BiCIKL as a basis for cross-cut research", "Industrial and commercial involvement"; JRA-01, JRA-05, NA-02, NA-03, TA, VA; Outcomes D4.2-4.3, D5.2-5.4, D6.1-6.4; Use case #2 in Suppl. material |
|
7 |
The integration of major scientific equipment and of knowledge-based resources (collections, archives, structured scientific information, RIs) leads to a better management of the data flow collected or produced by these facilities and resources. |
А key contribution of BiCIKL will be putting together open data integrations between RIs representing different data classes; the new system for discovery, validation and storage of bi- and multi-directional links, built in the project, will have a transformative impact on the way scientists address questions, generate their hypotheses and produce new knowledge. Research infrastructures are indeed known to be enablers of great scientific research, and it is difficult to anticipate the range of hypotheses and applications that will arise from the new data and knowledge arising as an outcome of the project. |
Number of research projects and publications using data linked across the entire BiCIKL community (for example: cyber catalogues).
|
Sections "BiCIKL contributing to the ESFRI Roadmap", "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "Transformed access for a new community of users", "Strengthening the European Research Area (ERA): Europe as a global leader in open data", Suppl. material Outcomes D4.2-4.3. D6.3-6.4; Use cases in Suppl. material |
|
8 |
The integrated and harmonised access to resourc es at EU level facilitate the use beyond research and contribute to evidence-based policy making. |
Beyond research, BiCIKL will add a significant value in serving policy decisions, e.g. combined use of data from collections, taxonomy and literature, can provide evidence on biological invasions, or historical dynamics of biodiversity and ecosystems, hence modelling and supporting informed policy decisions to combat environmental challenges. The CETAF will use its advocacy and engagement programs, specially through the European Initiatives Advisory Group, to outreach the EU Commission and countries governmental authorities, notably the NCPs. The CETAF strategy as a whole towards the participation of scientists in biodiversity assessments (CBD, IPBES), the public authorities outreach and advocating program through the Stakeholders Forum envisaged under DiSSCo, initiatives as clusters (ENVRI-FAIR) and future missions on Biodiversity will drive the efforts for the future. The public entities will be facilitated in their work by adequate tools for monitoring (dashboards, assessment tools, indicators) towards the big challenges (climate action, green deal, biodiversity loss but equally FAIRness, cloud repository and access, Earth digital, etc.) while private sector might be engaged through innovation and procurement. |
Worked examples of project outputs being fed to and used by the policy sphere. Policy briefs published by the project.
|
Sections "BiCIKL contributing to the ESFRI Roadmap", "BiCIKL - a cornerstone in the global Alliance for Biodiversity Knowledge process", "The BiCIKL community in a ten-years perspective", "Gender Balance and geographical distribution underpins BiCIKL", "Strengthening the European Research Area (ERA): Europe as a global leader in open data", "Communication activities"; NA-02-03, WP12; Outcomes D3.3-3.4, D9.1. |
BiCIKL focuses on promoting a trans-disciplinary approach to data use, along the entire data life cycle. By fully adopting FAIR principles and translating them into actionable policies across the network of partners, BiCIKL will put the interlinked FAIR data at the heart of data-driven scientific practices beyond domain limits.
To that end, the BiCIKL access programme will be instrumental. Based on a strong JRA and NA efforts which will introduce and propagate novel, formerly unparalleled, access services, BiCIKL will balance access modes to respond to the scientific needs of wider communities of practice. As such, it will prioritise content generation based on urgent scientific needs and provide unlimited, open and free VA to relevant data.
Strengthening the European Research Area (ERA): Europe as a global leader in open data
A recent Communication of the European Commission gives an EU perspective “to become a leading role model for a society empowered by data to make better decisions – in business and the public sector” with the aim by 2030 for the EU “to create a single European data space – a genuine single market for data, open to data from across the world”. BiCIKL is improving data, data infrastructures, interoperability of data and researchers capacity to work with big data, including the practically untackled and formerly unavailable and unlinked structured data from literature. All these activities and products will have strategic and economic importance to Europe and for other regions, since biodiversity is critically important anywhere in the world. The progress of digitisation of literature and collections and delivery of complex, cross-disciplinary datasets provides opportunities for new services which create employment, are inclusive, and have global reach. Digitised data can be integrated with other data far beyond biodiversity and genomics per se, allowing for the linking of expertise from different domains, and fostering the creation of new capabilities, as required by EU economic policies.
BiCIKL will directly contribute by delivering components of a much needed pan-European ecosystem of interlinked RIs along the ESFRI Roadmap, refining and harmonising data management practices across the participating organisations and promoting FAIR open access to biodiversity-linked information. By investing in virtual multi-modal access to data and by developing added-value scientific services that allow researchers to further benefit from the data availability, BiCIKL will contribute to the development of an urgently needed online pool of services that significantly lower the entry barrier for new and diverse users to create, access and interpret biodiversity-related data and thus, produce new knowledge and new research paths that can support advance of science in Europe but also worldwide.
During the BiCIKL project, a series of use cases will highlight the feasibility of combining data across disciplines and fields of science in order to advance scientific knowledge in areas relevant to urgent challenges. These new datasets created on the fly as a result of federated querying across domains will be used in fields as disparate as human health, invasive species, biodiversity loss and urban greening, with impact on a similarly wide range of end users.
Novel virtual access services and support structures
BiCIKL builds on the long established trust relationships between service providers and users to develop capacity enhancement, training modules and unified support mechanisms. Such mechanisms will be handed over to CETAF, DiSCCo, GBIF and LIFEWATCH to ensure their persistence. The project employs innovative ways of creating new and open scientific content through interlinking activities that are focused and synchronised across the data domains. By putting scientific needs at the centre of how the consortium generates new information from VA, and remote TA through open calls that are independently prioritised on their scientific merit and urgency, we will ensure the most impactful modalities of data re-use are identified for further improvement and development.
To fully optimise access to European RIs, it is imperative that biodiversity-related data domains are not disconnected modes, but rather complementary, providing more holistic ways of retrieving information from European scientific assets. To make this possible, BiCIKL will develop a novel service, providing a unified (one-stop entry point) to data and services through the BKH serving as a knowledge broker. The introduction of BKH will significantly lower overheads for both users and RIs’ administrators. Post-project operation of BKH will be ensured by the GBIF organisation that will host the access platform and by the rest of RIs that will promote and channel its further use. Working with the consortium on highly specialised aspects of work will allow SME partners to innovate in new areas with a variety of institutions, improving their competitiveness in these and other areas, such as software for distribution modelling or data extraction, collation or mining (
Policies, processes and standards harmonisation across organisations
To fully benefit from the technology and science innovations of BiCIKL, existing strategies and protocols for sustainable data annotation, storage, availability and analysis will be improved. Common standards are urgently needed to develop shared documentation policies and interoperable workflows. BiCIKL is introducing complementary work packages to address the gaps in existing standards around data sharing (NA-01) and ensure their improvement, testing and implementation (JRA, NA-02, NA-03) and use (TA and VA). These specifically address the minimum metadata sharing requirements so that to provide stable and meaningful linking between data records at the different RIs. Most of the BiCIKL activities across its three pillars will develop and implement a more complete corpus of standards to be incorporated into organisational policies and practices.
Barriers and other factors affecting the achievement of project objectives
The RIs participating in the starting BiCIKL community are aware that the main difficulties on the road to an integrated pan-European ecosystem of RIs is the fragmentation and insufficient interoperability at the level of data standards and access. Nonetheless, the work in previous EU projects supported several biodiversity RIs to reach the critical tipping points where cross-disciplinary data management and re-use for research and other purposes is already possible (example for such an approach of using data from different domains and RIs is the BIOPOLYSURF project). This obvious but still unrealised opportunity is exactly what BiCIKL will address by involving most advanced RIs in the different data domains to come together following and implementing a minimum of FAIR data sharing and linking standards, allowing the use of citation networks, from specimens and samples to literature where these are used, and afterwards linked back from the publications to the respective database records.
The BiCIKL partners are also aware by their past experience that too optimistic reliance on ‘community-maintained’ services and curation effort does not always ensure sustainability and long-term enhancement, hence a carefully thought set of incentives will be introduced to assure provenance record and credit to contributors and users. In addition to that, the new access services will generally focus on the ‘per-purpose’ incentives, rather than on the community willingness to curate and enrich data.
The COVID-19 outbreak happened during the writing of this proposal, hence the BiCIKL consortium is prepared for the various obstacles that the pandemic will cause to all aspects of economic and social life in Europe and globally. If necessary the project will be performed entirely online, without physical meetings of any kind. Should this happen, the travel budgets will be re-allocated by agreement between the partners and in coordination with EC, to support additional TA open call projects. Increasing efforts in community engagement and communication will also be put in place in order to mitigate the possible loss of physical meetings. Use cases and open call projects that study the zoonotic role of biodiversity (e.g. of pangolins and bats) will be seeked, encouraged and supported. One such use case (#2) is described in Suppl. material
Measures to maximise impact
Dissemination and exploitation of results
Overall approach
Continuous and effective dissemination of project results towards their potential user groups is key to realising and monitoring the expected impacts of the project. While dissemination deals with the targeted knowledge and information transfer to stakeholders who can potentially be users of the project results and products, exploitation deals with the concrete plans on the use of services and tools and ensuring their sustainability (
Another key component of the dissemination and exploitation activities will be a user engagement strategy to:
- develop BiCIKL starting community so that it meets user needs, and
- expand the user base (T1.1 in NA-01 and all activities through NA-02 and NA-03).
It will consider, for example, how best to work with users so that they can play a role in testing and developing new services. The aim will be to establish a fluent and fruitful relationship with users that can better ensure the sustainability of the BiCIKL community. It will take into consideration the fact that, in many instances, the providers of BiCIKL services and products are also (at times) users of those services (for example, a site manager may both provide and use data).
BiCIKL will address the full range of relevant end users and groups first via the project’s website and then via the BiCIKL Biodiversity Knowledge Hub (NA-02-03, T2.2, T3.1 and T3.3.). This platform will provide a dynamic support and instrumental means for communication and dissemination both in and outside the BiCIKL community. It will facilitate dissemination of project outputs by creating a hub to publish and promote project reports, scientific papers, policy briefs, press releases with linkages to other web-enabled communication channels and data sources (e.g. partner, actor group, or EU project websites, etc.). The platform will also be used to link to open source developments within BiCIKL, and to advertise for open source contributions from stakeholders (both internal and external).
Special emphasis will be placed on using existing international and national networks of taxonomists and museum curators (e.g. Taxacom and ICZN lists), as well as linking to on-going EU projects (SYNTHESYS+), large international conservation associations (IUCN, BirdLife International, WWF) and research networks (CETAF). This approach will enable BiCIKL to exploit new and alternative communication channels and opportunities, by actively seeking agreements with relevant actor groups to attend online or physical events, public meetings as well as to use their communication channels.
A series of dissemination and training events tailored to the needs of the different stakeholder groups are planned within BiCIKL (Pillar 1 NA) to ensure knowledge transfer and capacity building, including: data hackathons designed to establish best practices for creating and sharing biodiversity-related data (NA-01); a training programme on making the most of the Biodiversity Knowledge Hub (BKH) and BiCIKL services (NA-02).
A further objective of NA-03 is to establish - from the core of existing stakeholder and users - a solid user and supporting community, which can, with time, be further developed and moreover serve as a pilot for similar initiatives across domains elsewhere. A process to engage new members to the consortium will be established and will be made attractive by clearly highlighting the added value of the RI for the stakeholders.
Defining users and stakeholders
The resulting new European starting community of key RIs and its related services will be of interest and use to a wide range of stakeholders, including: researchers and research institutions, citizen scientists, data aggregators, public authorities, repositories, libraries, environmental regulators, and publishers. The primary user groups of BiCIKL services are taxonomists, molecular biologists, ecologists and biodiversity researchers in general, but apart from the academic community, BiCIKL will communicate with a far wider audience, including (but not limited to) data managers, librarians, private sector companies, etc. This preliminary analysis of the stakeholders and potential target groups will be further elaborated in the Revision of the Plan for the Dissemination and Exploitation of Project Results, based on findings and recommendations from an in-depth analysis of the stakeholder landscape, including identifying and classifying current and potential BiCIKL users, their needs and requirements (defined within Task 1.1). Profiles will be created for each resulting target group which will identify the groups’ preferable ways to receive information, relevant key messages and appropriate level of language.
Dissemination channels
BiCIKL will tailor the use of various uni- and bi-directional dissemination channels to the needs of each defined potential user group. The preliminary mapping of dissemination channels, target groups, related impact and relevant KPIs for which they will be applied can be seen in Table
Overview of the BiCIKL dissemination activities. Target audience key: Researchers (R); Decision Makers (DM); National Authorities (N); EU Authorities (E); Interested Organisations (O); Industry (I). Impact reference numbers (section "Transformed access for a new community of users") refer to: 1. Establish impacts; 2. Improve awareness and trust; 3. Harmonised datasets; 4. Improved monitoring; 5. Lasting cooperation.
|
Tool & Objective and contribution to project impacts |
Target Audience and (Impact) |
Monitoring tools and impact indicators (KPIs) |
|
Open source/access policy for all BiCIKL tool development to promote trust. Publication of open-source collaborative calls to engage external actors in JRA and tool developments. |
R, E, I (1,2,5) |
Google, YouTube, Facebook and Twitter analytics. Statistics for website traffic, page visits, link requests and downloads etc. Number of downloads, subscribers, users, communication exchanges and emails. KPI: 25 |
|
Public online library - Provide open access to papers, reports and deliverables, with linkages to journals and data repositories. |
R, O, I, N, E (1,2,5) |
|
|
Email alerts - Automated dissemination of news and announcements e.g. direct targeting of EC and other institutions for BiCIKL news. |
R, E, O, I (1, 2, 5) |
% average increase in website and social media traffic per year. |
|
Internal and external communication platform - Means to exchange information and discuss specific topics of common interest. |
R, N, O, I (2,5) |
|
|
BiCIKL presence at key conferences, such as TDWG, IUCN, CIBK3, BHL and others. BiCIKL presence at expert meetings and workshops, such as Biodiversity Summit 2021, 20th International Botanical Congress, Genome Engineering and Synthetic Biology, etc. just to mention a few examples. |
R, E, N, I (1-5) |
Meeting attendance. KPI: BiCIKL presentations/posters at BIS(TDWG) conferences (minimum 3 per conference) and CETAF and other meetings (minimum 1 per meeting). |
|
Workshops targeting dissemination and feedback from key stakeholder groups: MS3.4 - Workshop with RIs MS3.5 - Workshop with publishers. |
R, DM, I, O (1, 2, 5) |
Meeting attendance, meeting minutes. KPI: Two workshops, minimum 10 participants per workshop. |
|
Scientific publications - presentation of research findings and evaluation of its scientific quality through feedback from the scientific community in terms of open access, peer-reviewed publications. Special issues. |
R, E, N, I (1-5) |
Number & impact of publications. KPI: minimum 1 scientific publication in a peer-reviewed journal per TA project; minimum 3 publications or project partners presenting the key results of BiCIKL. |
|
Training on the use of the Biodiversity Knowledge Hub (BKH) and related services envisaged in WP2 (NA-02). |
E, N, I (1, 2, 5) |
Number of trainees. Downloads of modules. |
|
Production and distribution of specialised materials targeting the specific needs of each group (factsheets, infographics, policy briefs). |
R, O, I, N, E (1 - 5) |
Number of materials & downloads. KPI: Two training sessions with a minimum of 25 participants from the CETAF network of institutions and other stakeholders. Number of downloads of training materials at least 3 times higher than the number of trainees. |
|
Direct communication of key results to newsletters, bulletins, websites, blogs of key organisations operating in the sphere of interest of BiCIKL, including partners communities (specially through DiSSCo, CETAF, GBIF, LIFEWATCH and ELIXIR), relevant EU DGs (DG Environment), the global audience (e.g. via iDigBio in USA) and to a multidisciplinar target group (e.g. via BHL for heritage-related publications). |
R, O, I, N, E (1 - 5) |
Number of contributions. KPI: One project flyer, 5 factsheets and infographics describing the services developed in JRA-01-05, 2 flyers describing BKH and FDP, 2 policy briefs. |
All project dissemination activities will be further enhanced by the communication measures described in part 2.2.2 of this section.
Dissemination responsibilities
WP NA-03 “Implementation, stakeholder engagement and outreach for the Biodiversity Knowledge Hub” will be devoted to dissemination and sharing of knowledge. Furthermore, dissemination activities will be facilitated through the other WPs to utilise and extend the reach of BiCIKL dissemination using channels and networks of partners and other groups collaborating in the BiCIKL project. All BiCIKL partners will be actively engaged in the dissemination process by:
- Providing content to the communication team.
- Using their own personal and/or institutional networks, social media and websites to promote the project.
- Using relevant conferences to present the project results and distribute dissemination materials.
- Publishing research and data papers in reputable international scientific journals, in line with their academic and institutional policies.
- Participating in campaigns and events specifically designed as to raise visibility of the new community and increase engagement from relevant actors beyond the project consortium.
Existing large networks of interest groups that are indirectly involved in the project via partners (specially through DiSSCo, CETAF, GBIF, LIFEWATCH and ELIXIR) and their related initiatives will allow wide dissemination, reaching out to a global audience.
Open data strategy
The Horizon 2020 work program highlights the need to have research data openly used, by maximizing access to and re-use of these data. To coordinate data management within the project, BiCIKL will develop a guiding Data Management Plan (DMP) (D12.9). The DMP will specifically cover: handling of research data during and after the project; data collection and processing; methodologies and standards; data sharing and open access; curation and preservation. The DMP will also provide the dataset metadata specification that will be used in the data registry, following an appropriate relevant standard. It will specify the recommended licensing schemes, preferably using the Creative Commons Public Domain (CC0) and Attribution (CC BY) licenses as suggested by H2020. In the cases where the datasets cannot be publicly shared, the reasons will be mentioned in its metadata description (e.g. ethical, rules of personal data, intellectual property, commercial, privacy-related, security-related). Below is a preliminary description of all major points to be covered in detail within the project DMP:
- What types of data will the project generate/collect? Numerous and varied data sets will be collected or generated by BiCIKL project partners, e.g. data extracted from literature (traits, biotic interactions, people, institutions, projects and environmental relationships), taxonomic data (specimens, sequences, names, biological samples, specimen records); molecular data. The project will not only openly share data but will provide a unique new level of linking open data that will open the full research cycle and will encourage re-use and reproducibility along the line: specimens →sequences → species → analytics → publications → biodiversity knowledge graph → re-use.
- What standards will be used? To ensure interoperability, the BiCIKL project aims to collect and document the data in standardized formats (i.e. Darwin Core) to ensure that the datasets can be understood, interpreted and shared with accompanying metadata and documentation and relevant supporting material. Metadata standards will depend on the discipline and/or the methodology that was used to produce the data. BiCIKL partners will use discipline-specific repositories and common/standard metadata requirements and ontologies, e.g. TaxPub, OpenBiodiv-O, Ecological Metadata Language (EML), the latter being a metadata specification particularly developed for ecology and where necessary will be appended with other existing ISO-90155 compliant metadata libraries dependent on discipline-specific or institutional repositories.
- How will this data be exploited and/or shared/made accessible for verification and re-use? BiCIKL data will be openly shared through automated workflows with relevant repositories, including but not limited to the Biodiversity Literature Repository at Zenodo, Plazi TreatmentBank, GBIF, ENA, CoL, OpenBiodiv, PMC Europe.
- How will this data be curated and preserved? All data and models, both generated as part of BiCIKL and obtained from other sources, will be annotated, using internationally recognised keywords and meta-tags. Output from BiCIKL will be organised in an easily accessible and interpretable format. The necessary tools, standards and protocols for making BiCIKL data accessible, findable, exchangeable and secured on the long term will be made available to all BiCIKL partners and the BiCIKL users.
- Management of internal knowledge in BiCIKL. The terms of Intellectual Property Rights (IPR) management will be specified in detail in the Consortium Agreement to be signed at the beginning of the project. BiCIKL partners will work on a cooperative basis without commercial interest. However, for future maintenance of software, models and data mutual agreements on ownership and access conditions are essential to build trust and to respect interests relevant for durable cooperation. Issues about ownership, access rights and use conditions will be described transparently in the Consortium Agreement to ensure optimal cooperation among the BiCIKL partners. To that end, the Consortium Agreement will define use conditions. User groups, already foreseen in this project, will be asked to agree to these conditions using partnership agreements.
- Management of external knowledge in BiCIKL. Non-confidential results will be disseminated on the project website and through open access, i.e. free online access, applying the ‘gold’ open access model. Each WP leader has responsibility to manage external knowledge available to the general public according to the dissemination plan mentioned above. Fundamental scientific results will be freely disseminated through appropriate channels including scientific publications, presentations at international conferences and workshops. The publication venues will be primary scientific high-impact open access journals (such as the European Journal of Taxonomy (EJT), Biodiversity Data Journal (BDJ), Research Ideas and Outcomes (RIO), or other Pensoft’s journals). BiCIKL will follow the guidelines on open access to scientific publication and research data stated in H2020. Some budget for supporting publication in open access form will be dedicated and managed by the Executive Committee in accordance with the dissemination plan of the project and will also be available through the TA services of ARPHA-XML. Deliverables and other important project outputs will be published in a dedicated open access collection in RIO Journal.
- Open source policies. The software tools or plugins produced within the BiCIKL will be available as open source code under an appropriate license and published in the open science RIO Journal to ensure findability and reusability of all open source resources. The aim of BiCIKL’s open source approach is to ensure that a framework for new contributions is established that allows them to continue to be developed as open source in order to facilitate the further adoption and update of the technologies by all stakeholders.
Exploitation and sustainability of results
The exploitation and sustainability of the BiCIKL results and products assumes two levels of responsibilities:
- products and services developed at the base of either project partners or RI will be a responsibility of the respective partner or RI;
- the key synthetic products of BiCIKL, namely BKH and FDP, will be hosted and run after the project end by a large international organisation (GBIF) in the first case and a consortium of projects partners and RIs who provide services through FDP in the second case. The sustainability will be enforced by the uptake of the products and services by the starting community through actions and measures described in the Plan for the Dissemination and Exploitation of Project results (D3.2) and in the User engagement plan (Task 3.2.).
An essential element of the project sustainability is the adherence to the long term data preservation and accessibility via the repositories and RI involved (especially Zenodo, GBIF, ENA and actually all others) in compliance with the EOSC long term sustainability plans supported by the Member States and infrastructures. To ensure also the long term commitment to Open and FAIR data, BiCIKL will adopt whenever relevant the RDA FAIR Maturity KPIs to check the Fairness status of the data of the partners and infrastructures involved.
Communication activities
In addition to project dissemination, an active and strategically planned communication will support the project’s objectives through engaging society at large and providing information to interested persons. A communication strategy (M3.1) (following the platform designed in T3.1 and embedded into the more comprehensive Plan for Dissemination and Exploitation of Results (D3.2)), developed in NA-03 will outline the establishing of the BiCIKL brand and making effective use of a wide range of communication channels (including the BiCIKL website, newsletters and social media feeds).
Communications within the consortium, stakeholder advisory board and multi-actor forum: Communication and document exchange tools will be used as part of a web enabled communications and learning platform, to create a chat-like environment that simultaneously eases communication and streamlines information and access to documents into relevant channels to alleviate workload and stimulate fruitful and focused discussions. The BiCIKL communication and learning platform will have restricted space (intranet) for the consortium as well as participants engaged as part of BiCIKL’s stakeholder advisory board and multi-actor forum. It will provide a place where the BiCIKL project team and collaborators can communicate, share documents and work together.
External communication strategy: BiCIKL external communication strategies will be bi-directional, i.e. not only disseminating project outputs to targeted actor groups and the public at large, but also eliciting expertise, knowledge and perceptions as part of the project’s engagement activities. Social media will be used to create a community around the project and stimulate bi-directional communication and provision of feedback. Twitter in particular has proven to be an effective tool in the scientific community. A detailed social media strategy following the principles outlined in the recently published official EC Guidance: Social media guide for EU funded R&I projects (EC 2020). Press releases will be used to outreach to media at both international (Eurekalert!), EU and local levels to ensure important findings reach all corners of society and to instigate change in social perceptions. Specific press releases will be targeted at the EC and other interested policy bodies. Blogs and news will be posted regularly (minimum 1-2 per month) by the project partners and invited external parties. Multimedia clip(s) will be produced and used to promote key products or findings. All major communication channels to be used within BiCIKL are listed in Table
Overview of the BiCIKL communication activities. Industry (I). Impact reference numbers (section "Transformed access for a new community of users") refer to: 1. Establish impacts; 2. Improve awareness and trust; 3. Harmonised datasets; 4. Improved monitoring; 5. Lasting cooperation.
|
Tool & Objective and contribution to project impacts |
Target Audience and (Impact) |
Monitoring tools and impact indicators |
|
Project website - Inform and engage interested parties through provision of general project information, results, developments and outcomes with linkages to social media channels; promote increased media visibility to encourage network growth and focus on improved data availability. |
All (1,2,5) |
Google, YouTube, Facebook and Twitter analytics. Statistics for website traffic, page visits, link requests and downloads etc. Number of downloads, subscribers, users, communication exchanges and emails. KPI: 25 % average increase in web traffic per year. |
|
Provision of information (announcement of significant project results) about on-going events, project outcomes and related activ- ities tailored to targeted audiences by:
|
All (2 3, 4) |
Number of posts; number of re-tweets (Twitter); number of followers and “likes”; number of press release views; number of people who’ve watched and shared project videos. KPI: Two tweets per week from the BiCIKL Twitter profile and 3-5 retweets per week of posts related to the BiCIKL activities; 1 original tweet per week related to BiCIKL from each project partner’s Twitter account; 2 retweets per week related to BiCIKL activities from each project partner’s Twitter account; number of Twitter followers: 100 (first year), 250 (second year) 400 (third year); 25 % av- erage increase in likes and web statistics per year. |
Working together, dissemination and communication activities will support BiCIKL in achieving its expected im- pacts detailed in section 2.1. While dissemination will target the project’s stakeholders and potential users directly through supplying timely information about access to project tools, innovations, communication will ensure that the wider public is informed on the importance of the project’s activities towards achieving positive socio-economic impacts. Dissemination and exploitation activities in NA are designed to ensure sustainability and multiply the impact of improved and adequate usage of access services and tools under TA and VA and innovation under JRA. Project communication will not only inform society about the project but further support dissemination activities by multiplying messages and raising awareness through the communication channels toolset.
Implementation
Work plan
BiCIKL will consist of 12 work packages (WP) divided into three pillars, representing the three key activities: Networking Activities (NA), Joint Research Activities (JRA) and Access (Trans-national and Virtual Access, TA and VA, respectively). The relationships between the WPs involved in BiCIKL are illustrated on Fig.
Networking Activities (NA): Work packages 1-3 & 12. NA-01 (WP1), led by MBG, will ensure effective identification of current data needs, data standards and vocabularies, and the development of new or underspecified standards to make them suitable for data exchange and interoperability between involved infrastructures. NA-02 (WP2), led by GBIF, will deal with stakeholder mapping, build capacity for new partnerships, classify and arrange patterns of engagement flows between providers of data and provide training to users; NA-02 will also do the preparatory phase and lay the foundation for the Biodiversity Knowledge Hub (BKH), built in NA-03. Consortium-wide tasks on promotion, dissemination, communication, training and capacity building will be included in NA-03; NA-03, led by CETAF, will develop an information cluster and knowledge broker, the BKH, designed to promote and disseminate project tools, services and data provided by the new community. WP12, led by Pensoft, will manage the BiCIKL consortium, ensuring that the project’s tasks are coherently and effectively carried out both within the consortium and externally, and the effective implementation of the work packages on behalf of the EC, the project beneficiaries and the users.
Trans-national and Virtual Access (TA & VA): Work packages 4 & 5. Access to RIs, their associated expertise and specialised equipment is vital in the field of biodiversity research. TA (WP4, led by ELIXIR Hub) will enable remote trans-national access to data and services provided by partners to named users who have submitted a defined use case proposal via a project call process, which will strengthen the biodiversity expert’s capacity across Europe. VA (WP5, led by ELIXIR Hub) will involve nine research infrastructures offering virtual access to open FAIR data, tools and services for biodiver- sity researchers and other users, provided by the new community.
Joint Research Activities (JRA): Work pack- ages 6-10. Five JRA work packages will generate ground-breaking technological innovations to serve the user community. JRA-01 (WP6) led by Plazi will develop and implement novel technologies for next-generation publishing and enhance existing tools and create automated workflows for discovery, mapping, extraction semantic enhancement of FAIRified data from the literature. JRA-02 (WP7) led by Naturalis, will develop sustainable persistent identifier service for specimens and samples and link specimen records with other specimen-derived data available from the participating RIs, including scientific literature. JRA-03 (WP8) led by EMBL-EBI will build user tools and workflows that drive accurate and complete reporting of source annotations into molecular biology databases at time of deposition, and will establish networks of connected data from the data resources of molecular biology, natural history collections, taxonomy and literature. JRA-04 (WP10), led by Sp2000, will provide services for mapping significant alternative species classifications and will develop streamlined services to update Catalogue of Life backbone classification and link it to other data classes. JRA-05 (WP11), led by SIB, will deliver the analytical software tools needed to support the most advanced FAIR experience for members of the biodiversity community with a focus on Findability (search and question-answering) and Access to interlinked data, through the FAIR Data Place (FDP) interface.
Work package 1
WP1 - NA-01 Coordination and interoperability of infrastructures through harmonisation of community policies, standards and guidelines [Months: 1-18]
MeiseBG, NATURALIS, Plazi, ELIXIR/EMBL-EBI, CERN, SIB, UTARTU, LIFEWATCH, GBIF
Objectives
- Understanding user requirements from the community in terms of the questions they want to address with the data.
- Coordination of data standards usage between infrastructures, particularly exchange standards and controlled vocabularies.
- Development of new or underspecified standards to make them suitable for the needs of data exchange and interoperability between involved infrastructures.
- Alignment of data sharing policies and access procedures to facilitate easy use and transparency to the end user, whilst at the same time monitoring and reporting on use.
- Ensuring the data access methods, such as websites, downloads and APIs, conform to user requirements and optimum for the volume of requests they will receive.
Description of work and role of partner
Task 1.1. A technical user requirements analysis [Lead: LIFEWATCH, Partners: GBIF, MBG, ELIXIR Hub, Naturalis, SIB, Plazi, UTARTU].
This analysis will include users’ needs for types of data, quality needs and modes of access. There is a broad user-base for linked data and this must be reflected in best practice, sorts of data that are made accessible, the linkages between them and the modes of access to the different categories. On the one hand, there may be focused requests specific to single collection entities, such as finding all the specimens and sequences related to a publication. However, there are a whole range of requests, up to and including requests for the whole body of linked works, and further, requests for text and data mining across large corpora of text-based knowledge. The latter cases might include research into scientometrics. We will accommodate all levels of access and find modes of access that will suit user needs. For the starting community presented by this consortium, these user requirements will foresee future requirements going far beyond the status quo.
Task 1.2. Testing and streamlining interoperability [Lead: MBG, Partners: SIB, GBIF, ELIXIR Hub, Plazi, LIFEWATCH, Naturalis, TDWG].
Data can conform to the FAIR data principles at a number of levels and this is particularly true of interoperability where although systems can almost always be made to work together, this comes at a cost if not designed for it. This task will inform the infrastructures of their incompatibilities and ensure a level of interoperability sufficient for the user needs identified in the technical user requirements. We will pilot the use of combined infrastructure services through two hackathons of potential users to work on specific problems. That is, getting researchers and software engineers together to prototype applications of the infrastructure and test new ideas. These hackathons will be organised around themes to attract diverse user groups and use cases, particularly encouraging the participation of early career researchers. By actually using the integrated infrastructures in different situations we will be able to prove their potential and troubleshoot interoperability and access issues. We will report the success of these hackathons and the problems and solutions they uncovered in the process. Following on from this we will also specifically test the APIs and other means of access for interoperability for identified use cases. In conclusion, we will use the results of the hackathons to make specific recommendations to the infrastructures and to standards organizations to address the insufficiencies we uncover.
Task 1.3. Findability, reuse and accessibility alignment [Lead: Naturalis, Partners: GBIF, CERN, Plazi, LIFEWATCH, TDWG].
Data integration is as much about policy at an institutional and infrastructural level as it is about technical and semantic interoperability. The task will focus on (1) supporting the global adoption of agreed upon persistent identifiers for digital specimen objects, taxonomic treatments, collections, persons and taxa across stakeholders and in particular natural science museums (linked to Task 7.1). Within the data infrastructures, the task will document best practices for the generation, management and curation of the linked data. Also within the scientific publishing process, it will aim to harmonise data and literature publication policies (linked to Task 6.3). We will also look at how researchers are accessing infrastructures and whether this can be improved.
List of deliverables
- D1.1. User requirements analysis report (M12)
- D1.2. Recommendations for interoperability among infrastructures (M10)
- D1.3. Best practice manual for findability, re-use and accessibility of infrastructures (M18)
List of milestones
- MS1 The first BiCIKL data hackathon with partners and selected invitees has been held (M4)
Work package 2
WP2 - NA-02 Defining & co-designing the Biodiversity Knowledge Hub (BKH) and operational training [Months: 1-30]
GBIF, ELIXIR/EMBL-EBI, CETAF, LIFEWATCH
Objectives
- To map the landscape of community stakeholders across the biodiversity and life science domains, along with their functional roles, platforms, tools, services and practices, in order to cultivate opportunities for future innovation.
- To establish the building blocks and build capacity for new partnerships, including cross-organizational alignment, integrated approaches and coherent long-term governance.
- To classify and arrange patterns of engagement flows between providers of data and users of the derived services.
- To design the training required for full participation and collaboration of stakeholders in the broader portfolio of interlinked services at all stages, from data generation to analysis and application. WP2 aims to provide a more thorough picture of the roles, responsibilities and services of the literature-linked infrastructures and stakeholder organizations in the biodiversity and life science community. Stakeholder mapping of the relevant research infrastructures along with the tools, platforms and services serving stakeholders within the domain will enable a more robust assessment of approaches for building the Biodiversity Knowledge Hub (BKH). Having a well-informed working model of this multi-dimensional starting community will enable development of an effective operational training programme that can build capacity and address existing needs across the community.
Description of work and role of partners
Task 2.1. Identify and establish the necessary operational framework [Lead: GBIF, Partners: CETAF, LIFEWATCH].
This effort will catalogue research infrastructures within the biodiversity and life sciences domains. This mapping will describe interaction between the data providers and users that comprise our stakeholders. The effort will guide further partnerships needed to implement shared solutions that facilitate collaboration among the starting community. GBIF will lead the effort, under the auspices of the Alliance for biodiversity knowledge, a stakeholder-driven initiative to increase global collaboration and coordination across biodiversity informatics-related communities (
- identifying stakeholders, partners and research communities;
- cataloguing information about stakeholders;
- documenting their interest in the project’s linked outputs;
- engaging them as contributors to or users of the starting community’s infrastructures.
Characterizing the relationships will ensure that the BKH can integrate multiple sources of information from the collaborating infrastructures. The analysis will use an inclusive approach aimed at building trust and maximizing cooperation, helping to cultivate opportunities for future innovation by aligning with and engaging stakeholders that are now isolated.
Task 2.2. Identify the components of the Biodiversity Knowledge Hub [Lead: GBIF, Partners: CETAF, ELIXIR Hub, LIFEWATCH].
This task will build on Task 2.1 by further characterizing the functional roles, tools, services, and use cases. The analysis will map a network graph of functional relationships, describing:
- access requirements for different data types;
- constraints on data mining, linkage and usage;
- benchmarks for ensuring data quality standards, compliance with FAIR standards;
- key intervention points for deploying innovations;
- metrics that signal priorities for additional opportunities for adding new services.
This network graph will form the basis of the BKH and will integrate interlinked analytics services (WP10). The graph will help inform the development of content for Task 1.2, by refining operational flows; WP1, by characterizing system requirements; WP3, by shaping the structure of BiCIKL; JRA, by reflecting the users captured needs. Tasks 2.1 and 2.2 will build on similar efforts in both the EU-funded ICEDIG project and DiSSCo Prepare. GBIF and LIFEWATCH will convene the Third Global Biodiversity Informatics Conference (GBIC3), under the banner of the alliance, in 2021 (M2.1), leveraging the BiCIKL collaboration and bringing together outside stakeholders to review and provide input into the network graph.
Task 2.3. Translate the operational framework and functional network graph into an educational cloud [Lead: CETAF, Partners: GBIF, LIFEWATCH].
This task will focus on enabling community members to perform activities effectively. Materials will outline how users access biodiversity data interlinked through the published literature and between the data classes. Advanced publishing tools will enable users to retain access to the rich publication metadata and the semantic structure. This task will establish a web-based training programme, based on a training needs assessment, integrating training modules on data liberation and publishing tools (M2.4) and the best practices identified by Task 3.3. The training programme for capacity building will be delivered as a Massive Open Online Course (MOOC) (D2.3) with the following topics:
- Web-based structure and functioning
- Based on relational databases approach
- Based on Linked Open Data
- The BiCIKL website developed as the overall web-based structure for the project will also deliver the training programme.
- Community supporters and stakeholders in the scientific, publishing and educational domains
- Platforms used, including DEST, the CETAF European School for training
- Definition of the training programme, including:
- general on-line training (and supporting material) for generic sets of users, including the best practices and recommendations produced under T3.3
- specific modules (as e.g. on data liberation and publishing tools)
- customized training modules for specific subcategories of the targeted audience (e.g in the field of multilingual word embeddings (MWEs)
- Business models
- New semantically enhanced publishing workflows (CETAF-MNHN, ARPHA-XML) for authors and publishers as well as new publishing platforms’ users.
List of deliverables
- D2.1. Relational network graph for the Biodiversity Knowledge Hub (BKH) (M15)
- D2.2. Stakeholder input into the network graph provided by Third Global Biodiversity Informatics Conference (GBIC3) (M15)
- D2.3. BiCKL training programme MOOC (M30)
List of milestones
- MS2 Third Global Biodiversity Informatics Conference (GBIC3) (M15)
- MS3 Network graph database (M15)
- MS4 Workshop for training needs identification (M20)
- MS5 Training Modules on data liberation or publishing tools pilot activity (M29)
- MS6 Best Practices and recommendations (from T3.3) translated into training material (M29)
Work package 3
WP3 - NA-03 Implementation, stakeholder engagement and outreach for the Biodiversity Knowledge Hub [Months: 1-36]
CETAF, PENSOFT, MeiseBG, ELIXIR/EMBL-EBI, LIFEWATCH, GBIF
Objectives
- Ensuring a distinct identity of the project and its services through branding, visualisation and elaborated dissemination and communication strategy.
- Active dialogue between the key biodiversity-related research infrastructures in Europe with researchers and other stakeholders through a modern, multi-functional, communication and publishing platform.
- Promotion, dissemination and communication of the multi-access options provided by the new BiCIKL community.
- Elaboration of a one-stop portal for a Biodiversity Knowledge Hub, operating as the information cluster for access to all tools and services provided by the participating RI.
- Refinement and testing of alliance-based virtual infrastructure which will facilitate interactions among project RI and other international RI that can benefit from BiCIKL developments. WP3 will be focusing on facilitating access to research infrastructures by modern and technically advanced active dissemination and communication methods and ensure the sustainability of the project results through the Biodiversity Knowledge Hub (BKH), developed in the course of the project and hosted on the GBIF infrastructure after its expiration. This includes the engagement in EOSC through the deployment of a VRE taking advantage of existing Blockchain technologies.
Description of work and role of partners
Task 3.1. Community branding, website and communication platform [Lead: Pensoft, Partners: GBIF, CETAF].
Within the first months of the project, a recognisable project visual identity will be developed alongside an initial marketing pack (a project logo, brand manual, brochure, poster, letterhead and presentation templates). These will ensure that BiCIKL is communicated effectively and professionally with the objective to raise awareness and build a community from the very start. A Communication strategy (M3.1) embedded into the more comprehensive dissemination Plan for Dissemination and Exploitation of Project Results (D3.2) will set the rules and KPIs to guide and measure communication activities during the project’s duration. A modern and user-friendly public website will provide an easy-to-navigate, continuously updated platform allowing fast access to general information about BiCIKL and its activities, operating on several levels; internal communication system (described in more detail in Task 12.3), publishing platform for community-related documents (reports, standards, guidelines, policy briefs, training programme. etc), blog and public relations interface, social media profiles and others, to ensure effective integration, prioritization, cost effectiveness and sustainability of the community’s communication interface, networking activities and operations during the project lifetime and especially beyond it through the Biodiversity Knowledge Hub (BKH) (Task 3.3). The platform will be secured for rapid uptake and long-term persistence by bringing together and multiplying the strong brands of the participating research infrastructures, their extensive experience and existing communities. Social media profiles will be created and maintained in Twitter and Facebook with the goal to create an active and engaged online community around BiCIKL.
Task 3.2. Engagement and dissemination [Lead: CETAF, Partners: GBIF, Pensoft, ELIXIR Hub, LIFEWATCH, Allpartners].
On top of the BiCIKL Tool platform (T3.1), progress and achievements will be widely disseminated following the Plan for the Dissemination and Exploitation of Project Results (D3.2 due in M6). It will identify the best use of tools (newsletters, press releases, messaging, etc), the most effective channels (social media, events, etc.), and the target audiences. The Plan will highlight best practices both internally and externally. A revision of the Plan (M3.3 due inmonth 24) will provide a mid-term evaluation. The Plan will give special emphasis to engage with stakeholders and to attract new communities of users along the entire data life cycle. To that end, and in close relationship with the virtual resources developed under T3.3, a set of 4 experts round tables (for tackling Virtual Access field, WP5 and contributing to JRA activities (WP6, WP7, WP8, WP10 and WP11)) will be organized to collate expert advice from non-academic stakeholders, including industrial actors and to foster cross-disciplinary fertilisation (which outcomes will be gathered under a joint report M3.7). Additionally, two focussed presentations/workshops to targeted audiences (for RIs under WP2 and WP3 and for publishing-related actors under WP3 and WP4) will be organized (milestones M3.4 and M3.5). All these engagement mechanisms will work towards the Alliance for biodiversity knowledge (Alliance) (arising from the Second GBIC Conference in 2018), to facilitate collaboration across biodiversity infrastructures and communities. The BKH will further advance the Alliance`s approach with virtual workshops, curriculum, best practices and models for collaboration at all scales among others, in biodiversity informatics. The dissemination and engagement actions will envisage the publication of policy briefings, reports and guidelines also highly valuable for decision-makers, public actors, private entities and communities of practices beyond the scientific realm. The most relevant and impactful results will be gathered in a collection of articles, published in the Research Ideas and Outcomes (RIO) journal. It will equally reflect that the project follows at all instances the "open science" practices.
Task 3.3. Implementation of the BKH and outreach [Lead: LIFEWATCH, Partners: GBIF, CETAF, Pensoft, ELIXIR Hub].
BKH will serve as a one-stop location and knowledge broker for users to find information about all services, tools and data provided by the participating infrastructures, including direct links, explanatory pages, guidelines, helpdesk and technical support. The initial prototype (M3.2) will begin as a subpage of the project website before being launched as a standalone portal within the GBIF infrastructure, where it will be maintained after the project expiration. To that end, and in order to further develop this BKH prototype, several recommendations and best practices will be developed, at the same time this will be duly engaged in EOSC through a proper VRE developed by LIFEWATCH ERIC taking advantage of using LifeBlock e-infrastructure features. As defined above, Task 3.2 will apply these best practises and recommendations in stakeholder engagement, by means of implementing RTs and asynchronous online consultations, when required, to encourage wider global participation in BiCIKL activities by enabling the participation of those who are unable or cannot afford to travel to specialized meetings. Virtual engagement also promises to significantly reduce the project’s carbon footprint - an obvious goal at present for any biodiversity informatics community. The best practices and recommendations will be delivered (M3.6) as training material to guide the implementation of the BKH and detail an open science decision-making process for the end user. Those best practices will be embedded into the Training programme produced under Task 2.3.
List of deliverables
- D3.1. Project logo, marketing pack and website design and development (M3)
- D3.2. Plan for Dissemination and Exploitation of Project results (M6)
- D3.3. Outreach initiatives report (M36)
- D3.4. Тhe Biodiversity Knowledge Hub (BKH) (M24)
List of milestones
- MS7 Communication strategy (M6)
- MS8 Biodiversity Knowledge Hub prototype (M12)
- MS9 Dissemination plan revision (M24)
- MS10 Workshop with RIs (M24)
- MS11 Workshop with publishers (M28)
- MS12 Biodiversity Knowledge Hub Best Practices communicated to T2.3 (M29)
- MS13 Outreach activities report I (M12)
- MS14 Outreach activities report II (M24)
Work package 4
WP4 - TA-01 Trans-national access to biodiversity infrastructure and services [Months: 1-36]
ELIXIR/EMBL-EBI, MeiseBG, FUB-BGBM
Objectives
- Enable trans-national access to data and services provided by seven leading research infrastructures to named users who have submitted a defined use case via a project call process.
- Run a small pilot call for projects starting after the first year. Out of this call will be funded 2-3 projects involving a subset of the WP4 partners.
- Run a full project call, starting at the end of the second year of the project. Out of this call will be funded 6-8 projects, involving all of the WP4 partners.
- Attract users to the new community and help establish BiCIKL as an enabling function for Biodiversity specialists across Europe.
Description of work and role of partners
The BiCIKL project aims to establish a new Biodiversity focussed community that brings together experts from the museum, molecular, literature and taxonomic realms. The aim is to better support taxonomists, ecologists and environmental scientists as described in sections "The fragmentation of the biodiversity informatics and how BiCIKL will improve it", "The BiCIKL contribution to fostering a culture of international cooperation", "Improved access via BiCIKL as a basis for cross-cut research" and "Transformed access for a new community of users" of this proposal. During this establishment phase, significant work on the infrastructures is needed before users can be actively supported by the new community. A key activity of the BiCIKL community in the longer term (subject to funding) would be to run open project calls across the scientific community in Europe. Many of the proposals we would expect to be made in those project calls will involve functionality that is not currently supported by the individual infrastructures. As a consequence, we do not plan to run a full project call in BiCIKL until the end of the second year, when much of this preparatory work will have been completed. Once the BiCIKL community is established and more mature, it is expected that such project calls as described here, could be run on a routine basis, also after the project expiration, if funding were available, for instance via future funded EC projects. Alternatively, or in addition, the use cases that we address herein, could help define a broader Biodiversity technical infrastructure that would start to reduce the hands-on support burden that is required, again subject to available funding.
Beneficiaries of WP4 - Modality of access: Work Package 4 is at the core of what the new BiCIKL community will produce, namely a new service that supports efficient integration of diverse data sets to meet the needs of user groups of named scientists with an interest in Biodiversity, from across Europe and beyond (up to 20 % of TA will be provided to research teams of prevailing third-country users). The key feature of TA is the provision of services to user groups from country(-ies) different from the host country of the RI installation which could be provided through remote transnational (no visits needed) access to data from at least two of the participating RIs requested by a user group. Physical access in BiCIKL will be possible but not financially supported by the open call projects; the successful applicants will have to cover their travel and subsistence costs from other sources, for example through synergies with the SYNTHESYS+ project. Task 4.4 will hold the majority of the budget in WP 4 which will be distributed on an as needs basis, defined by the projects that are selected and agreed by the infrastructure leads in-line with the unit costs for each infrastructure.
Support offered under this proposal: The specific support provided to users will vary depending on the infrastructure and is described individually below. Broadly speaking, it is foreseen that support will include tasks such as: assisting with access to data (for instance via large or complex queries) or specimens, data management and archiving or support with the use of complex bioinformatics tools (sequence assembly, clustering and annotation), publications using semantically interlinked data between several RIs, and automatic annotation pipelines for full text articles with semantic search engine.
Project eligibility: Defining the precise project eligibility for the two calls will be carried out as part of Task 4.1, 4.2 and 4.3 described below. However, there are three key criteria that will apply to both calls, and that can be defined in advance:
- Project proposals must concern a topic of relevance to the BiCIKL Community as a whole and should address relevant scientific questions, which could focus on taxonomical, ecological, environmental and other related topics.
- Project proposals must include at least two of the named infrastructures, where each infrastructure is focused on a different discipline (Museum specimens, Sequences, Taxonomies, Literature).
- Proposals must show a clear indication to publish any findings in an open access relevant journal, using new methods of structured publishing developed within BiCIKL.
Project call process: The project call process will be defined and executed by the lead beneficiary of WP4, EMBL via the ELIXIR Hub. There will be two calls: A pilot call to identify 2-3 projects for the community and a full call to identify 6-8 projects, for completion by the end of the BiCIKL project. The specific details of the call process will be fully defined in Task 4.1 (see below) but in summary, is envisaged to run as follows:
Define proposal eligibility criteria applicable to that call → Announce call via BiCIKL website and other channels → Open call for submissions → Close call and evaluate submissions for eligibility → Conduct external scientific reviews → Hold internal review panel to select projects → Determine how the projects will be delivered → Allocate project funds to infrastructures to execute projects → Run projects and review output → Evaluate project impact and effectiveness of project.
We will ensure there is no possibility of discrimination (by gender, race or religion or of any type) or bias within the call and review process, which will be run in as transparent any open way possible, subject to the usual need to protect the confidentiality of applicants and reviewers. The Equality and diversity champion appointed in the project will participate in all evaluation processes to ensure gender and other equality issues are properly considered. To establish and run this process, which is planned to be used in future more mature iterations of the BiCIKL community, will require the following tasks:
Task 4.1. Define the process for how the project calls for the users of the BiCIKL community will be run [Lead: ELIXIR Hub, Partners: Pensoft, Plazi, SIB, EMBL-EBI, MBG, BGBM-FUB].
This task will define how project calls will be made in the BiCIKL community, including the pilot and full phases. The Task lead will take input from the other WP4 partners and the executive leadership before finalising a process that will be published as a milestone via the BiCIKL website. Key elements of the process that will be defined will include: How call eligibility criteria will be agreed, timing and process for each call; how proposals will be reviewed and successful projects selected; how the funds for the project will be allocated to project partners and finally; how impact of the projects will be identified and disseminated. The final process will be described in a report and published via the BiCIKL website.
Task 4.2. Run the pilot phase project call [Lead: ELIXIR Hub].
At the end of year one (M12), and applying the process as defined in Task 4.1, a pilot project call will be run, with narrow eligibility criteria with the aim to identify 2 or 3 project proposals for consideration by the BiCIKL community. The details of the eligibility criteria will be defined once BiCIKL has started, but for the pilot phase are likely to be informed by the capabilities of the infrastructures at that time, and thus may not include all infrastructures. Specifically, the pilot phase will allow the BiCIKL community to:
- Support a small number of users with aim to create better defined user stories, which will support future activities of the BiCIKL community.
- Pilot the call process prior to releasing it to a wider set of potential users (via wider eligibility criteria).
- Pilot the mechanisms the community will need to distribute funds equitably across the various infrastructures to deliver on individual use case proposals.
- To start to inform BiCIKL of the details of the types of proposal that may be received. Given this is a new Community, this is an important aspect of the Pilot call.
Task 4.3. Run the full project call process [Lead: ELIXIR Hub].
At the end of year two and applying the process as defined in Task 4.1, and learning from the pilot phase, a full project call will be run, with broader eligibility criteria. The aim will be to identify 6-8 project proposals for consideration by the BiCIKL community. It is anticipated that the project proposals that will be made could be diverse and variable in nature. For instance, proposals could focus on needs to primarily integrate data from museum specimens and molecular data repositories. Alternatively, projects may focus on integrating literature and observational data. This won’t be known until the project proposals received are evaluated. Specifically, the full phase will allow the BiCIKL community to:
- Support a larger number of users, with more diverse requirements, with the aim to highlight a number of use cases where access to integrated biodiversity data provides demonstrable improvements over the current model and brings scientific benefit.
- Fully establish the call process, helping to define the longer-term sustainable model whereby this community could continue to operate.
Task 4.4. Management and distribution of BiCIKL funds to infrastructures to support trans-national access via project calls [Lead: ELIXIR Hub].
A key challenge for the BiCIKL community will be to establish robust mechanisms for how funds that will support the Trans-national access to the infrastructures will be distributed to the WP4 beneficiaries. The process to support this element will be defined in Task 4.1. Task 4.4 will hold the budget that will be distributed to the BiCIKL beneficiaries in support of the project call proposals.
Task 4.5. Formulate and report a long term plan for how the service offered by the BiCIKL community could be offered in the future [Lead: ELIXIR Hub].
A key output for WP4 will be an increased understanding of the need and the impact of offering services that link the diverse infrastructures including the likely impact of access to more systematic biodiversity-related knowledge corpora could have on broader Biodiversity goals. A report will be written and published via an appropriate external channel that describes the overall need and impact.
List of deliverables
- D4.1. Project call definition and execution plan, published via BiCIKL website (M3)
- D4.2. Report on pilot call process (M24)
- D4.3. Report on full call process (M36)
- D4.5. Report on a long term sustainability plan for the BiCIKL community services (M36)
List of milestones
- MS15 Project call process definition phase completed (M3)
- MS16 Pilot call projects process opens (M12)
- MS17 Full call projects process opens (M24)
Work package 5
WP5 - VA-01Virtual access to biodiversity infrastructure and services [Months: 1-36]
ELIXIR/EMBL-EBI, PENSOFT, NATURALIS, Plazi, CERN, UTARTU, LIFEWATCH, GBIF, sp2000
Objectives
The new BiCIKL community will include nine infrastructures that offer virtual access to any user in the Biodiversity community. Continued virtual access to all infrastructures will be required through the life cycle of the project to support Biodiversity-related research for scientists across the world. WP5 will:
- Provide helpdesk support and technical assistance for free and open access to any user to the tools and services provided by the new community.
- Ensure that users of the services can continue to access relevant data and services for the duration of the project.
Description of work and role of partners
Task 5.1. Coordination of virtual access across the named infrastructures [Lead: ELIXIR Hub, Partners: GBIF, Naturalis, Plazi, Pensoft, LIFEWATCH, CERN, SP2000, UTARTU].
The BiCiKL project will enable access to nine different infrastructures, providing very diverse types of service to any user worldwide. These infrastructures, individually, are key repositories of critical data for Biodiversity scientists. Collectively they provide a network of interconnected sources of data that allow users to address complex questions in the field. As the project progresses, the ease with which those questions can be addressed will gradually improve, for instance as the technical improvements described in the JRA components of the project are implemented. To ensure that access to the services by the wider Biodiversity community is maintained, to coordinate virtual access across the infrastructures and to monitor how the improvements to the infrastructures are impacting users we will pursue two subtasks:
Sub-task 5.1.1. Annual review of the BiCIKL infrastructures. To be held at the same time as the BiCIKL annual general assembly (or Kick-off meeting, as appropriate), an annual review will be held of the nine infrastructures that offer virtual access to include current status, changes since the previous review, overall funding landscape and future plans. This analysis will be included in the annual report.
Sub-task 5.1.2. Summaries of each infrastructure, their tools and services. Summaries of the annual reviews with the associated analyses and recommendations will be prepared and hosted on the BiCIKL website to serve as a landing site for new users.
Modality of access under this proposal: Access to the participating RIs will be given through their usual access procedure.
Support offered under this proposal: Each RI will appoint a contact person for the coordination of VA activities within their institution(s) either through a helpdesk or via a single point of contact for users. VA coordinators will act as a hub to coordinate VA requests and link them to appropriate specialists across the institution, to develop innovative proposals that are both practical and conform to the requirements of the VA programme. Most of the participating RIs manage their own data portals for the access to readily available material in a straightforward and user-friendly way. RI contact persons will offer support to users for the full range of services offered by the RI under BiCIKL project.
As well as general helpdesk support, specific services will be offered and widely advertised at some RIs. For example, at relevant conferences and via social media communication channels.
Many RIs already have considerable experience with supporting smaller on demand projects and have qualified managerial staff and data managers, who will engage in the BiCIKL project as required.
Outreach to new users: The EU-funding of BiCIKL will allow the RIs participating in this WP to reach out to researchers beyond their usual market and in a context of providing more integrated services. Opportunities, as they arise, will be taken to use relevant international conferences to reach out to new users.
Review procedure under this proposal: The BiCIKL VA access process guarantees a broad distribution of the VA-possibilities and fair treatment of the requests. The number of web-hits to data portals will be monitored and results analysed to demonstrate impact. VA coordinators will assist in collating statistics and metrics for VA assessment (including specimens digitised, demand versus supply, rate of download / resulting outputs, dataset citations, citations of papers in which data are used). The infrastructures will be periodically reviewed by the Access Provision Panel. Prior to the board meeting, the statistics and metrics gathered from each infrastructure will be collated and summarised into a single report document, which will be provided to the board in advance of the meeting. In addition, guidance will be provided to the board regarding how the statistics and metrics can be interpreted, building on the experience that ELIXIR has gained in selecting and monitoring the ELIXIR Core Data Resources.
List of deliverables
- D5.1. World class virtual access to the infrastructures in support of Biodiversity and related subject matter research is provided (M36)
- D5.2. Periodic assessments of VA by external board and a final assessment report (M36)
List of milestones
- MS18 Initial teleconference between infrastructures and follow-on regular TCs will have been established and set up (M3)
- MS19 External board review received I (M12)
- MS20 External board review received II (M24)
- MS21 External board review received III (M36)
Work package 6
WP6 - JRA-01 Liberation of data from literature, next-generation semantic publishing and delivery of FAIR data [Months: 1-36]
Plazi, PENSOFT, MeiseBG, ELIXIR/EMBL-EBI, CERN, CETAF, SIB
Objectives
- Develop and implement novel technologies and workflows for next-generation prospective publishing of structured content and data.
- Enhance existing tools and create automated workflow for discovery, mapping, extraction, semantic enhancement, curation and delivery of data liberated from PDF and XML literature sources.
- Align the use of identifiers and bi-directional linking of FAIRified biodiversity data between literature and RIs (including journal publishers) to enable distributed systems to interface with one another.
- Improve and extend automated workflows for seamless interoperation between knowledge, information and data through RDF-conversion of published content into LOD resources.
Our rationale for JRA-01 is to more rapidly extract, expose and interlink information in the published literature both after publishing (legacy literature) and during (rather than after) the publication process (prospectively published literature). We will Create and Extract (Tasks 6.1, 6.2), Recognise (Tasks 6.1 and 6.2), Identify (Task 6.3), Interlink (Tasks 6.2 and 6.4.) and Deliver (Tasks 6.3, 6.4) data throughout two parallel workflows, namely:
- data liberated and enhanced from legacy literature through text and data mining, and
- an advanced, prospective publishing process (Fig.
5 ).
This will be achieved by leveraging community standards and Linked Open Data principles.
Description of work and role of partners
Task 6.1. Develop novel publishing tools and workflows for structured, standards-aligned content [Lead: Pensoft, Partners: Plazi, EMBL-EBI].
Specifically, this task will apply innovative data science solutions to improve open access authoring and publishing. It will integrate narrative and data in an efficient and highly automated way, interfacing with the data life cycle at all stages: from data providers, through peer-reviewed and data-audited publications, to the FAIR data pool (including conversion to Linked Open Data in Таsк 6.4). Through these steps, we will accelerate the generation of new knowledge with state-of-the-art information technologies. We will thus take a definitive step towards resolving the “PDF impediment” to knowledge sharing through facilitating data-driven and semantically enriched publishing, based on both generic and domain-specific standards and ontologies. The task will evolve the existing ARPHA Writing Tool (AWT) into an independent authoring and editing environment to allow automated import, export and submission of both standardized data and metadata through use of community standards and conventions, including MinSEQE, MIxS, Darwin Core, DwC-A, BIOM, and semantic resources such as the Trait Ontology, ENVO, and GloBI. The AWT will generate (TaxPub) JATS XML documents. These augmented documents can be used for
- submission to journal publishing workflows, or
- computational workflows for efficient harvesting and further re-use of semantically enriched content.
Task 6.2. Liberation, enhancement, management and distribution of data from literature [Lead: Plazi, Partners: Pensoft, CERN].
This task will provide the tools to scale up and automate the workflow converting scholarly publications from both legacy literature and present-day publishers (Task 6.1) into structured data, which includes the text flow, document structure, and generic elements such as figures, captions, tables, bibliographic references and in-text citations, following three main stages:
- Automated workflow for article import from three formats (XML, digitally-born PDFs, scanned PDFs) using an improved version of the Lycophron and Refindit reconcile service tools to clean up metadata and to discover the existence of DOIs for batch import of articles into the BLR. A triage system will be developed to decide whether a document based on scans will be converted using OCR or double keying via a vendor. For XML-input, the requirement for a minimal specificity of JATS or TEI will be defined.
- Enhanced tools and services for data liberation and access will be assessed and further developed to yet automate the conversion workflow that processes the documents to a generic level of markup including the establishment of the text flow, structural elements like figures, tables, their respective caption, and bibliographic references, as well as their respective citations in the text.
- Domain-specific semantic markup tools, defined by the user requirement in NA-01 will be enhanced to cover named entities, taxonomic treatments and their structure, materials citations and sequence accession numbers, specimen and collection codes, using international formats (JATS) combined with domain-specific annotations (TaxPub, LOD, stable identifiers). This workflow can also be used to fit the often small-scale, autonomous journals who need to make the transition from PDF-only publishing to semantically enhanced XML.
Task 6.3. Federated FAIR data system through aligned identifiers [Lead: Plazi, Partners: Pensoft, CETAF, CETAFMNHN,SIB, CERN, MBG].
The existing tools used to FAIRify data (e.g. taxonomic treatments, figures, tables) and their metadata will be reviewed for compliance with the standards aligned in NA-01 and adjusted accordingly. Lookups for automated linking (e.g. accession codes, specimen codes, taxonomic names, person names) will be developed including a mechanism to notify the target RI of data not yet present in their holdings (e.g. such as GRSciColl or CoL). Tool and the API at BLR/Zenodo to provide bi-directional linking of citations (including batch operations) will be developed in coordination with Task 7.4 (specimens), Task 8.1 (sequences) and Task 10.4 (taxonomic names). The existing quality control tools for the extracted data will be assessed, further developed, and more tightly integrated into the production workflow. A workbench for curation of the data and links between data records will be developed in a collaboration with SIB (Task 11.5) to enable a wider community to curate extracted data, such as material citations or taxonomic names. The output formats will be augmented by the requirements formulated by the BiCIKL community (NA-01).
Task 6.4. Integrating published narrative and Linked Open Data [Lead: Pensoft, Partners: Plazi, SIB].
This task will use the existing OpenBiodiv-O ontology, which re-uses and interoperates with many community ontologies (e.g. SPAR-based ontologies), to enhance the interoperability of all content generated and interlinked by Task 6.1-6.3 through automated RDF conversion to Linked Open Data. Vitally, the associated OpenBiodiv system will integrate both legacy and prospectively published literature, creating a uniquely valuable LOD-based knowledge graph for biodiversity science. The access to data will be facilitated through novel applications designed to satisfy the most widely relevant competency questions raised by biodiversity scientists (NA-01). FAIR-ness (especially interoperability) will be ensured through multi-ontology mapping of literature corpora in JRA-05. The SIB Literature services (JRA-05) will further merge BLR (Task 6.3), and OpenBiodiv contents into EuropePMC via the SciLite gateway.
List of deliverables
- D6.1. Enhanced, standalone and freely accessible installation of the ARPHA Writing Tool (AWT) (M24)
- D6.2. Automated workflow for discovery, semantic enhancement, mapping, extraction, FAIRifying, curation, and dissemination of data liberated from literature (M36)
- D6.3. A workflow for bi-directional linking of data from literature to external resources (M18)
- D6.4. Applications for interoperable access to OpenBiodiv through semantically enhanced queries (M20)
List of milestones
- MS22 Specification of the independent authoring and editing environment (M6)
- MS23 Initial scoping and assessment of optimal degree of automation for processing workflow (M9)
- MS24 Prototype implementation of proposed advanced automation tools (M21)
- MS25 Beta-implementation and deployment of automated data extraction and preparation service (M30)
- MS26 Scoping and specifications for workbench (M12)
- MS27 App specifications (M10)
Work package 7
WP7 - JRA-02 Providing core access services and FAIR data on specimens and samples [Months: 3-36]
NATURALIS, Plazi, MeiseBG, ELIXIR/EMBL-EBI, UTARTU, FUB-BGBM
Objectives
This work package proposes to develop core access services for harnessing the power of specimen and sample data to advance research and making them FAIR research resources for the members of the biodiversity and other disciplines.
- Design and development of a sustainable persistent identifier (PID) service for specimens and samples.
- Ensure infrastructure links between specimen records and other specimen-derived data.
- Use Wikidata to support entity recognition links to specimen records.
- Serve FAIR specimen records.
Specimens stored by the world’s natural science museums and botanic gardens and other material samples stored by various organizations provide valuable resources for research. The current ecosystem of managing data derived from specimens and samples in Europe and globally is highly fragmented across disciplinary communities and stakeholders including museums, academic institutions and individual researchers. A multitude of institutional catalogues, diverse practices for sample identification, and discipline-specific data and metadata standards exist. Thus it is important that the data harvested and derived from these specimens and samples can be delivered in a FAIR way that is widely-adopted, inter-disciplinary and flexible.
JRA-02 is needed to create a persistent identifier registration, discovery, resolution and indexing service so that the data related to the specimens and samples can be unambiguously retrieved, re-used, linked, published and cited.
JRA-02 will leverage the current knowledge around persistent identifier implementation in various research infrastructures and also align with partners in Task 6.3 to ensure FAIR data access along the full research data lifecycle.
Description of work and role of partners
Task 7.1. Rolling out a pan-European PID system for Digital Specimens [Lead: Naturalis, Partners: BGBM, MBG].
A pan-European persistent identifier (PID) system will support the creation of identifiers for the digital representation of specimens and samples (Digital Specimens or DS) ensuring that the identifiers are globally unique, persistent and resolvable. This system will also manage associated metadata, facilitate tracking of provenance, enable discovery, link to other derived data and digital content. As part of the requirement gathering and design tasks, longterm sustainability and robustness need to be assessed. This system should support an estimated 30 billion identifiers and must be resilient for 30 years or more, and potentially beyond 100 years. A special focus will be on harvesting semantically enriched “Linked Open Collection Data” generated locally by distributed collection curators. These links, for example for persons or geographic entities, open up collection data as an integrated information space with a high potential for advanced interdisciplinary applications. The need for Persistent Identification of the digital representation of specimens in cyberspace does not only have European dimensions. It effectively touches upon a global need for consensus on digital specimens identification. During the Task we will facilitate a global conversation for reaching consensus, on the basis of the relevant BiCIKL outputs. Finally, further work will be done towards the ratification of a relevant open data standard (Open Digital Specimen -OpenDS), through the Biodiversity Information Standards Organisation (TDWG).
Task 7.2. Establishing functional links between specimens and other data classes [Lead: BGBM-FUB, Partners: EMBL-EBI,UTARTU, MBG, Plazi].
Over the past two decades massive amounts of DNA sequence data have been generated and uploaded to public DNA databases (e.g. INSDC and BOLD). The current recommendations for referencing specimens and samples using, for example, the GBIF Triple-ID are not sufficient to link data in a secure and machine-readable way. This task will perform a current FAIR assessment and coordinate with JRA-03 to establish links between specimens and derived sequence data. We will document and publish the current procedures for referencing collection objects and DNA samples in sequence data and vice versa (with WP8). Following on from this, an improved workflow and the required services to support the linking of sequence data with persistent specimen identifiers during submission will be specified. The system will search for suitable registrations in the existing specimen registration systems (e.g. CETAF, DiSSCo, GBIF) based on the available submission data and propose them for linking. For collections that want to enrich their databases with sequence data, a service will be implemented to search the INSDC databases for records matching existing specimen data. The technical implementations will be flanked by improved recommendations and best practices for linking specimens and samples with sequences that have been agreed upon in the community. Finally, we will provide a system to track metrics of usage and compliance with recommendations in INSDC data. The methods and workflows for enriching specimen data with links to sequence data citing specimens will be extended to “other data classes” such as literature (link to WP6).
Task 7.3. Using Wikidata to link and validate entities discovered in specimens [Lead: MBG, Partners: Naturalis, BGBM-FUB, Plazi].
The growing availability and complexity of specimen data urges us to use novel approaches to reduce complexity and to guarantee sustainability, transparency and interoperability. Through the establishment and improvement of linking algorithms, we will link collection-based data to a much broader body of knowledge about the world by leveraging the Wikidata infrastructure. We will use Wikidata as an information broker to provide stable identifiers for entities that do not exist in other data stores and as a bridge between different databases. We can also benefit from the human-in-the-loop aspect of Wikidata, where a global pool of editors will contribute to an ever richer source of data. In this task we will provide cycles of data analysis and enrichment that will successively improve the quality and usefulness of the data as the project progresses. Analysis of these linked data will inform us of many aspects of our collections that will valorize them and support better curation. For example, we will link taxonomic authors of species names to stable identifiers. We will link specimen collectors and determiners to stable identifiers where they can be uniquely identified. These people will be disambiguated using biographical resources from Wikidata and from those data in the collection databases of MBG, Naturalis and BGBM. People, their identities and biographical details, are one of the most important entities of the biodiversity knowledge graph. Identities of people connect taxa to publications, specimens and organizations. The availability of citable bibliographic details make person information an important source for the cross validation of data together with specimen and publication data. Furthermore, Wikidata entries contain links to other identifiers such as ORCID ID, taxonomic author abbreviations, International Standard Name Identifier and the Virtual International Authority File (VIAF).
Task 7.4. Providing specimen information as FAIR Digital Objects [Lead: Naturalis, Partners: BGBM, MBG, UTARTU].
Building on the work of Tasks 7.1-7.3, as well as the DiSSCo RI Data Management Plan this Task will focus on delivering key components that support the FAIR Digital Object Architecture of DiSSCo. FAIR Digital Objects (FDO) are atomic and stable over time, are uniquely and globally referenceable and accessed by the DO Interfacing Protocol. All relevant metadata information is included so that computer agents know what they are, what they can do with it and whether they are allowed to re-use their content. FDOs are serviced by trustworthy repositories using a wide variety of technologies and can be bundled to complex collections also being FDOs. They contain any type of digital information and its type can be associated with operations enabling automation of workflows and efficient data science. In this FAIR Digital Objects approach for specimens (
List of deliverables
- D7.1. Architecture Design for a pan-European PID system for Digital Specimens (M18)
- D7.2. Data standards and workflows for linking sequence data with specimens implemented and published (M30)
- D7.3. A repeatable informatics workflow that links people and places to stable identifiers of those entities, including a validation and preservation step (M30)
- D7.4. Digital Object Interface Protocol (DOIP) enabled Digital Object repository installation to store and provide digital specimen information (M36)
List of milestones
- MS28 Architecture design ready for evaluation by potential end users; design and specification report (M9)
- MS29 PID system development roadmap and technical provisioning for test system (beta/pre-production) (M14)
- MS30 Draft recommendation for specimen links in sequence data (M24)
- MS31 Full data and API specification for specimen links in sequence data (M27)
- MS32 The design and prototype of a workflow integrating Wikidata into validation and linking (M25)
- MS33 Documentation on how to interface with FAIR Digital Object via Digital Object Interface Protocol (DOIP) (M27)
Work package 8
WP8 - JRA-03 A data foundation for connected molecular, natural history collections and taxonomic data [Months: 1-24]
ELIXIR/EMBL-EBI, PENSOFT, Plazi, MeiseBG, UTARTU, FUB-BGBM, sp2000
Objectives
The methods and tools developed in BiCIKL will establish networks of connected data from the data resources of molecular biology, natural history collections, taxonomy and literature. In order to maximise the richness, and, hence, utility, of these networks, a foundation of well-structured and accessible annotations in the molecular biology databases is required. In the European Nucleotide Archive (ENA), the ELIXIR Core Data Resource in which primary nucleic acid sequence data are maintained, there are many annotations that relate to the sample source for an organism, for instance a natural history collection; a biobank; or a culture collection. While such annotations exist for a great many sequence records, they are either incomplete (many sequences are not linked to their sources), ambiguous (many annotations lead to multiple endpoints relating to the source), nor necessarily accurate (many incorrect annotations exist). In this work package, our objectives are:
- To establish a foundational layer of data for connected molecular, natural history collections and taxonomic data through at-scale curation, organisation and structuring of molecular biology data and services.
- To build user tools and workflows that drive accurate and complete reporting of source annotations into molecular biology databases at time of deposition.
- To facilitate data update cycles as inaccuracies and omissions come to light.
- To enable structured publication of molecular ecology data insufficiently served by current infrastructure.
Description of work and role of partners
Task 8.1. Profiling of existing source annotations, at-scale curation and data services [Lead: EMBL-EBI, Partners:ELIXIR Hub, UTARTU, BGBM-FUB, MBG, Plazi].
We will profile the current systems in use to capture and present source annotations, including the “specimen_voucher”, “bio_material” and “culture_collection” tags that are loosely captured in a Darwin Core-like structure. We will infer the identity of institutions from institution codes (free-text in the current data model) that are provided and map to a single normalised dictionary, using such context as voucher identifiers and taxonomic names as well as by querying institution collection databases where these exist. We will look more deeply into collection and voucher codes (again, free-text in the current model) and establish curation rules that disambiguate and correct them such that they can map to source materials. Applying this knowledge to the entire database through automated processes, we will provide a single, harmonised annotation that more directly and more accurately leads to the appropriate source material endpoint. We will track and respond to pan-European PID developments in Тask 7.1, applying these where and when available and contribute input data for Тask 7.2. Finally, we will provide an API to allow data to be discovered, queried and retrieved based on the single, harmonised annotation, which will be used, inter alia, as a basis for the tools to support collection curation in task 7.2.
Task 8.2. Data submissions tools and user workflows to capture improved sequenced material source annotations at point of deposition [Lead: EMBL-EBI, Partners: ELIXIR-HUB, UTARTU].
Source material annotations are submitted to ENA at the time of data deposition or at subsequent update. With many routes to submission (such as web interfaces, RESTful APIs and locally installed command-line submission tools, there is no single point into which tools to support accurate submissions can be integrated; rather there are many. We will build open-source software tools that will be downloadable and executable across platforms that will provide validation functions, allowing users preparing their data submission accurately to structure sequence material source annotations. Supplementing this with a clear how-to guide (that we will submit for publication likely as an “application note”), we will encourage the use of this tool amongst communities submitting relevant data. Finally, we will integrate the tool where appropriate into our submission systems and supplement with additional features (such as auto-complete options and pull-down menus to capture accurate institution codes in web tools).
Task 8.3. Operation of the ELIXIR Contextual Data Clearinghouse for the reporting of source material annotation gaps and inaccuracies [Lead: UTARTU, Partners: EMBL-EBI, Sp2000, ELIXIR-Hub].
Our ELIXIR Contextual Data Clearinghouse serves to allow those with improved or corrected annotations, such as sequenced material information and sequencing library details, to feed this information back to primary repositories. Offering light and simple transactions across a RESTful API, the current system enables those with such annotations (such as secondary databases that consume and curate data from repositories) to report at will - in a single batch operation or as individual annotations emerge. The data repositories (such as ENA) then access the staged annotations and effect updates as appropriate for their operations. The decoupled nature of the system reduces complexity both for annotation provider and recipient repository and thus encourages greater reporting. We will build a web interface for the ELIXIR Contextual Data Clearinghouse with focus on the reporting of errors and gaps in sequenced material source annotations. The API and web interface will be promoted to those communities (such as taxonomists, those abstracting from the literature - within and beyond BiCIKL - and natural history collections managers) with the appropriate knowledge and tools to feed enhanced annotations detailing such information as material sources (e.g. through PID schemes developed under Task 7.1) and taxonomic names (e.g. from Task 10.3) and literature (from Task 6.3).
Task 8.4. Creation of a data repository, with submission and access tools, for molecular ecology data [Lead: EMBLEBI; Partners: ELIXIR Hub, UTARTU, Pensoft].
A gap in molecular databases exists in ecological and other environmental applications of molecular biology in that there is no structured data repository for ecological and other matrix data that are derived from sequences and provide such information as occurrences and abundances of genes and species. While previous work under the ELIXIR Marine Metagenomics Community called for the deposition of such information (
List of deliverables
- D8.1. Availability via API of single, harmonised sequenced material source annotations in ENA (M20)
- D8.2. Validation tool open-source software release (M18)
- D8.3. Web interface for ELIXIR Contextual Data Clearinghouse (M9)
- D8.4. First submission and access tools available for BIOM data (M24)
List of milestones
- MS34 Report on profile of sequenced material source annotations (M6)
- MS35 Submission for publication of “application note” relating to sequenced material source validation tool (M14)
- MS36 First improved sequenced material source annotations routed through the ELIXIR Contextual Data Clearinghouse (M6)
Work package 9
WP9 - Ethics requirements [Months: 1-36]
PENSOFT
Objectives
The objective is to ensure compliance with the 'ethics requirements' set out in this work package.
Description of work and role of partners
This work package sets out the 'ethics requirements' that the project must comply with.
List of ddeliverables
- D9.1 : POPD - Requirement No. 1 (M1)
Work package 10
WP10 - JRA-04 Delivering a trusted and evolving taxonomic framework for data integration [Months: 2-30]
sp2000, MeiseBG, ELIXIR/EMBL-EBI, CETAF, UTARTU, GBIF
Objectives
- Encapsulate taxonomic understanding for interpretation of scientific names and other species identifiers (including MOTU identifiers from BOLD and UNITE) within other components of BiCIKL and by the wider biodiversity information user base.
- Support FAIR access through standardised identifiers, metadata and version histories for names, MOTUs and other taxon concepts.
- Develop robust approaches to handling and presenting alternative classification schemes.
- Develop tools and workflows to enable taxonomic experts rapidly to review, validate and improve handling of all names and identifiers, particularly those inserted into the classification via automated processing.
Taxonomy plays a central role in managing biodiversity information. This affects almost all user interactions, either as a primary filter or as fundamental information for interpreting results. Taxonomic knowledge is essential for navigating through a linked open data graph of biodiversity knowledge. JRA-04 is necessary as a result of the complexity and dynamic nature of research into species and their classification. New species are described daily. Older species concepts are revised. Understanding of evolutionary relationships changes. Molecular research probes difficult or megadiverse groups and offers new systems to map their diversity, often suggesting alternative relationships. Important data link to names and concepts, but accessing these data depends on access to the best current understanding of their inter relationships.
JRA-04 focuses on FAIR improvements to all stages in the process to harvest, aggregate and curate taxonomic information from publications and genomic research. This includes accelerating standardised access to streams of new or digitised species treatments and MOTU classifications, mapping of all of these into a consolidated Catalogue of Life (CoL) (initially dependent on many decisions from automated processes), offering initially automated mappings between CoL and other significant species checklists, enabling expert community curation of all these derived products, and encapsulating these in services and visualisations that support the infrastructures of all BiCIKL partners and other users.
Description of work and role of partners
Task 10.1. Automated ingestion of published taxonomic information [Lead: GBIF, Partners: Sp2000, UTARTU].
Structured taxonomic treatments published in new publications (under WP 6) or mined by BLR from BHL (Biodiversity Heritage Library) and other legacy publications will be automatically ingested into the Catalogue of Life as collections of species treatments. MOTU classifications from UNITE and iBOL (BOLD BINs) will also be streamlined as compatible collections of treatments. All of these collections will be published to the Catalogue of Life Clearinghouse and stored as FAIR datasets, with version histories where applicable.
Task 10.2. Data mapping for taxonomic information [Lead: GBIF, Partners: Sp2000, EMBL-EBI].
The Catalogue of Life will be enhanced to provide a continuously improving classification constructed from best available components for each taxon and augmented with names from other sources (e.g. GBIF Taxonomic backbone, NCBI Taxonomy) all components being available for curation and validation using tools to be developed through Task 10.3. Other species checklists published to the CoL Clearinghouse will be mapped against the Catalogue of Life classification to facilitate access and reinterpretation of data using alternative schemes (with key regulatory and national lists prioritised). These mappings will also be open for curation and validation using Task 10.3 tools. When new versions of contributing datasets are published, these mappings will be repeated, including use of any previous curatorial decisions. MOTUs will be presented as rankless children of the lowest-ranked inclusive taxon. A workflow, based on Refindit reconciliation service tools (Task 6.2) will be implemented to resolve publication references in species treatments to the appropriate publication entities (from JRA-01).
Task 10.3. Curation and validation of taxonomic information [Lead: Sp2000, Partners: UTARTU, GBIF, EMBL- EBI,MBG].
Workbench tools will be developed and community access models will be established to allow taxonomists and other knowledgeable curators to verify, reject or annotate prior decisions on the handling of names, concepts and MOTUs. Mechanisms will be developed for authorising the scope of user contributions and for handling and representing cases of disagreement. Curators will also have the ability to provide missing data elements and to validate and curate automated mapping between the Catalogue of Life and other species checklists. Incorrect assignments to molecular records will be reportable by these curators into the ELIXIR Contextual Data Clearinghouse (Task 8.3).
Task 10.4. Delivery and presentation of taxonomic information [Lead: Sp2000, Partners: GBIF, UTARTU].
User communities will be consulted on the content, structure and formats required for efficient use of taxonomic information and on improved search, browsing, presentation and visualisation of the Catalogue of Life and other checklists, including comparative visualisation and data mapping between checklists. Particular focus will be given to encapsulation of CoL services in forms that can be used directly throughout the infrastructures of BiCIKL partners to provide consistent and predictable search and discovery behaviours for all taxonomic information.
List of deliverables
- D10.1. New and updated taxonomic data products automatically visible in taxonomic framework (M30)
- D10.2. Workflow for bi-directional linking of taxonomic references to literature (M24)
- D10.3.Validation and curation workbench for taxonomic data (M18)
- D10.4. Web visualisation of mappings between CoL and other checklists (M12)
- D10.5. CoL services for direct encapsulated use within search and discovery services of BiCIKL partner infrastructures (M27)
List of milestones
- MS37 Specification for curation workbench (M12)
- MS38 Implementation of workbench released for use by taxonomic users (M21)
- MS39 Specification for encapsulated services (M18)
- MS40 Integration of encapsulated service into first partner infrastructure (M27)
- MS41 Implementation of mapping visualisation released (M27)
Work package 11
WP11 - JRA-05 FAIR Data Place: linking, finding and access [Months: 3-32]
SIB, PENSOFT, NATURALIS, Plazi, MeiseBG, ELIXIR/EMBL-EBI
Objectives
This package aims to deliver the analytical software tools needed to support the most advanced FAIR experience for members of the biodiversity community with a focus on Findability (search and question-answering) and Access (linking). These services will fulfil the following key objectives:
- Discover, display and store bi- or multi-directional links between the four main domains: specimens, sequences, taxon names/OTUs and biodiversity literature.
- Develop and provide advanced services for question-based search and answering, across the different domains.
- Design and implement a workbench through Interactive Graphic User Interfaces to support the curation of multi-domain linking, searching and question-answering.
JRA-05 will leverage different resources. Firstly, it will use the wealth of semantically enriched literature, automatically annotated with named-entities (species names, accession numbers, and taxonomic features and anatomical traits, including measurements, colors, etc.), as delivered by JRA01-04. Thus, direct literature-based cross-references (e.g. MEDLINE, PMC) using accession numbers linked to curated molecular biology databases (e.g. ENA, UniProt) and specimens resources (e.g. GBIF, DiSSCo, GGBN), will be complemented by inferred links. Secondly, the research activities will capitalize on AI analytical resources. All assertion links generated and stored (e.g. DOI, PMID, ENA Accession Numbers, etc.) will be exposed via a documented public API in compliance with FAIR and virtual access principles.
Description of work and role of partners
Task 11.1. Benchmarking collection [Lead: Plazi, Partners: SIB, ELIXIR Hub, Naturalis].
Derived from NA-01 user requirements, a search and question-answering evaluation collection will be developed to assess the search and question-answering effectiveness of the project’s FAIR Data Place (FDP). The document collection shall include:
- A broad-coverage list of biodiversity resources, including specimens catalogues, literature annotated with onto-terminologies (e.g OntoBee, NCBI Taxonomy, Uberon and others, as appropriate) as explored in WP6;
- A prioritized list of factoid questions (#25-100 following TREC guidelines) and;
- The associated relevance judgements to capture community-specific information requests.
Cross domain questions could be:
- What is the body size or an organism with sequence X?
- What is the color pattern of the species of a genus?
- Are color patterns linked to a geographic pattern and genes?
The resulting benchmark will follow the Cranfield paradigm and will be made publicly available to the community. The generation of the relevance judgements, i.e. the associations between a question and a relevant document/passage will be based on curated databases and the comparison with general purpose web search engines.
Task 11.2. Distributed search, link prediction and validation [Lead: Naturalis, Partners: SIB, ELIXIR Hub, MBG].
We will develop an user-friendly interface to allow end users to efficiently validate existing and newly predicted links. The biodiversity resources defined in Task 11.1 will be either harvested or federated with the Open-BioDiv knowledge graph (Task 6.4) system in order to obtain a seamless integration between them. The knowledge graph and associated entities will be converted to a format suitable for (deep) learning algorithms using recent graph embedding techniques. Link prediction algorithms for knowledge graphs will be used to visualize new links between various entities, e.g. database accession numbers, literature citations, as well as specimens and taxonomic relationships. Entities and links will be displayed through an intuitive interface that allows switching between the visual representation of the original knowledge graph (links, entities) and the embedded representation. This allows us to efficiently identify complex meaningful connections in a knowledge graph consisting of millions of facts. Active machine learning techniques will be used to efficiently curate entities and links by showing the ones predicted as most uncertain to the user first. Fully automatic link generation will be performed on a limited subset of entities when confidence scores will reach a given precision level (e.g. 95%).
Task 11.3. Passage retrieval [Lead: SIB, Partners: Plazi, Naturalis, Pensoft].
We will develop a document/passage retriever to retrieve multiple passages for each question. This will be a ranking model that utilizes the representations of questions and passages to compute their relevance and to assign a confidence score to each passage. We will rely on the latest advances in question answering and reading comprehension, such as neural character-level models, language models, self-attention recurrent networks for representing questions and passages, which will be combined with traditional vector space models, such as Okapi BM25. Information found in external knowledge resources can be incorporated in embedding representations and also can be used to increase training data for our machine learning approaches. Tasks 11.3 and 11.4 will benefit from the same open data and software stack. Machine learning frameworks (e.g., PyTorch, Scikit-learn) will be used to build various association models, to discover new facts and generate new hypotheses out of a growing corpus of liberated data. Regarding data, the JRA-05 will benefit from supervised machine reading and question-answering methodologies and from the multitude of large-scale datasets, such as Wikipedia (e.g. WikiReading, SQuAD) and general web sources (e.g. MS MARCO, TriviaQA, SearchQA). These general datasets will be combined with domain-specific resources (e.g. colors, measurements, origin) to build an interactive curation-support platform for the community.
Task 11.4. Factoid question-answering [Lead: SIB, Partners: Naturalis, Pensoft].
Capitalizing on the document/passage retrieval service, we will generate a domain-specific language model: state of the art methods will be implemented (e.g. BERT, XLNet) and cross-model experiments will be performed to derive a robust biodiversity resource. Several training datasets will be tested to evolve a system able to answer biodiversity-related questions (taxonomic treatments, extraction of taxonomic relationships, etc). A collection of factoid questions (see Task 11.1) will be used to assess the effectiveness of the system with priority given to factoid questions, whose answers can be relatively cheaply obtained either fully automatically or with limited labour force. TREC-like evaluations will be performed with priority to be given to high precision (e.g. Mean Reciprocal Rank) to complement mean average precision measures needed for Task 11.3.
Task 11.5. Search and Question-Answering Portal [Lead: SIB, Partners: Plazi, Pensoft, Naturalis].
The JSON services described in this JRA will be integrated within an interaction-rich user interface powered with responsive web technologies (HTML 5.5, Javascript, Angular). Via this Workbench, end-users will be able to screen and curate through the ranked or classified results to perform triage/answer extraction tasks. The interaction will be recorded in order to provide a continuous evaluation of the accuracy of the services (e.g. TREC measures) and ultimately to streamline the iterative learning of the respective models. The design and integration of the interface will be achieved using Agile methodologies and Rapid Application Development with the active participation of the end users. The tracking of questions leading to conflicting answers yet validated by the different users of the system via web analytics (e.g. click through) will make possible the identification of inconsistencies in the universe of the platform. The system will benefit from the crowd curation functionalities developed in a collaboration with Task 6.3.
List of deliverables
- D11.1. Evaluation benchmark: a database of questions associated with a set of relevant articles (e.g. PMIDs) to measure the progress of the WP towards an effective search service (M12)
- D11.2. Search and link association services: A RESTful API, which will input a link/accession number and return a ranked list of neighbors links with a confidence score (M18)
- D11.3. Passage retrieval services: A RESTful API, which will input a set of keywords and return a set of articles/passages ranked by relevance (M18)
- D11.4. Factoid question-answering: A RESTful API, which will input a factoid question in natural language and extract a short answer out of the articles/passages returned by D11.3 (M24)
- D11.5. Workbench implementation: a set of responsive Graphic User Interfaces developed in HTML 5.x/JavaScript to operate the aforementioned services. (M32)
List of milestones
- MS42 Gathering of a limited set of biodiversity-related questions (M6)
- MS43 Mockups to support the design of the link association services (M15)
- MS44 Mockups to support the design of the workbench (M28)
Work package 12
WP12 - Project management [Months: 1-36]
PENSOFT, NATURALIS, MeiseBG, ELIXIR/EMBL-EBI
Objectives
Professional and permanent project management is crucial for a successful and effective execution and management of the project. Therefore, WP12 is designated as an on-going activity over the full project duration, in order to provide the methods, tools and infrastructure to manage the project efficiently; to facilitate internal communication and collaboration and gear the team towards the fulfillment of the project objectives and contractual commitments. The key objectives are:
- Ensure that project’s deliverables are produced on time, in high quality and within budget.
- Provide an effective interface with the Contracting Authority.
- Organise and maintain smooth internal communication on all aspects of the project such as project progress, decisions, documents and version control, resource use and deliverables.
- Timely plan, organise, support the workflows and follow up on all governance and management events of the project.
- Oversee the quality control measures and ascertain risk.
- Execute robust financial management and timely reporting.
- Ensure project activities and service content conform to ethics and data protection principles.
- Keep expenses, material use and travel-related emissions to a minimum and maximise the smart use of technology and talent shared by the Consortium.
Description of work and role of partners
The project Coordinator Pensoft bears the full responsibility for all contractual arrangements and reporting to the European Commission and the overall success of the project. The Coordinator`s project management role will be supported by the in-house Project Management Team (PMT), employed by Pensoft, who will be in charge of the daily tasks of coordination, administration, finance and internal communication. For detailed description of the management bodies and their basic operational principles please refer to Section "Consortium as a whole".
Task 12.1. Detailed project planning and Standard Operational Procedures (SOP) [Lead: Pensoft, Partners: Naturalis, MBG, ELIXIR Hub; All partners in the General Assembly Meetings; CETAF, GBIF, Plazi, SIB and Species 2000 in the Executive Board].
During the first three months of the project Pensoft will draft and present to the Consortium and the Contracting Authority the detailed Project Work Plan (D12.1) with the Standard Operational Procedures (SOP). This document will describe in detail the work plan, responsibilities, governance and project management structures and procedures, the roles and composition of the project management bodies, and a schedule for their operation. In parallel, the Project Management Team (PMT) will prepare the Inception Report (D12.2) for approval by the project partners (beneficiaries), and will communicate it to the Contracting Authority. Another communication to the Authority will cover the access cost units of the participating research infrastructures who have chosen the cost unit method. Since the consortium partners are committed to Open Science principles, a Data Management Plan (DMP) (D12.9) will be produced within the first 6 months of the project. This deliverable will evolve during the lifetime of the project in order to present the status of the project's reflections on data management. The format of the plan will follow the EC template provided on the Participants Portal. The Coordinator will also draft the Final Project Report which will be presented to the EB and GA for approval at M34 before finalisation at the end of the project.
Task 12.2 Organisation of General Assembly and Executive Board meetings [Lead: Pensoft, Partners: Naturalis, MBG, ELIXIR Hub].
BiCIKL brings together highly experienced project partners who collaborate bi-laterally among themselves as a matter of routine. Therefore, the organisation of project meetings will be kept to the necessary minimum to ensure the basic governance and exchange of interactions. The opportunity of face-to-face meetings at the annual General Assembly will also be used to run specific workshops and in-depth working meetings. The following meetings will be organised (in case of force majeure circumstances like the present COVID-19 pandemic, the Consortium will switch to online-only meetings):
- Annual General Assembly (GA): The GA events will bring together physically all project beneficiaries. The GA decisions will be mandatory for the entire project. The agenda will cover detailed planning of work packages and tasks, hackathons, cross-task technical workshops and communication events [M1, 18, 34] and will deliver a GA Report (D12.3-5). At the first kick off meeting, the EB will present the detailed work plan for the project and all partners will be thoroughly informed about their specific roles, tasks, responsibilities and rights deriving from the project work plan and the SOP. These two documents will be included in the comprehensive Project Inception Report (D12.2).
- Executive Board Meetings (EB): The EB meetings will take place every month, starting from M2 and will be mostly online through tele- and video conferencing. They will be summoned and chaired by the Coordinator. The agenda, minutes and action points of each meeting will be taken and kept in good order by a member of the PMT. The EB will give each WP leader the opportunity to review progress, coordinate timing and provide/receive feedback from the Coordinator, the Pillar leaders and the remaining WP leaders. The decisions taken by the EB will be mandatory for the Coordinator.
Task 12.3. Internal communication, document management and archiving [Lead: Pensoft, Partners: Naturalis, MBG, ELIXIR Hub, All partners].
The internal communication within the consortium is a key factor for the implementation and the success of the project, therefore, the Coordinator will put together specific effort in ensuring it is run professionally and effectively. Pensoft has recognised expertise in providing advanced IT platforms and solutions. An Internal Communication Platform (ICP) will be set up especially for the project (D12.6), based on successful applications in other projects. It will provide a password protected area of the project’s website featuring the following tools:
- Mailing module, collecting contact information of all partners and providing group mailing options for all key project nodes (different work packages, Executive Board, financial managers etc.);
- Internal project repository, delivering a safe and convenient storage for all project documents and files. The storage can be subdivided into different levels of access according to the project structure and responsibilities distribution and needs of the project. Protocols will be developed to ensure standardised file-naming and versioning.
Task 12.4. Administration and financial management and reporting [Lead: Pensoft, Partners: Naturalis, MBG, ELIXIR Hub, All partners].
The Coordinator will ensure that the contractual aspects within the Consortium are carried out in a timely manner, including partner accession to the contract, establishment of and compliance with the Consortium agreement, and fulfilment of the beneficiaries’ contractual obligations. It will implement appropriate quality control tools to monitor the work progress including deliverables and milestones, prepare and coordinate the periodic project reports with the partners, and ensure timely delivery to the Contracting Authority. The Coordinator will moderate potential difficulties and internal conflicts, propose resolutions and corrective actions. The Coordinator will also be responsible for the financial and administrative management and efficient use of the resources and manage the distribution of the Community financial contribution between the contractors and activities in accordance with the contract and the decisions taken by the Consortium (AG and/or EB).
Task 12.5. Risk management and quality control [Lead: Pensoft, Partners: Naturalis, MBG, ELIXIR Hub].
Risk management and quality assurance are two of the key responsibilities of the project Coordinator, and they will be provided through the following mechanisms:
- Risk management: By directly and closely overseeing the potential contingencies that could affect the expected progress (in terms both of time and quality) the Coordinator/ PMT will forecast, detect and assess any potential risks. Risks or issues that require closer scrutiny, as well as risks that are subject to monitoring since they have been pre-identified, will be reported to the Executive Board (D12.7, D12.8). The agenda of the regular (monthly) meetings of the EB will cover a risk management review and the EB will determine any necessary follow-up action, subsequently reported to the ensuing General Assembly. The GA is the annual forum for wider discussion and course correction of project tasks and activities in case of important risks would require amending.
The PMT will provide a helpdesk for day-to-day queries from partners on contractual and financial issues relevant to the Project.
- Quality control: Quality control will be ensured through peer review of Deliverables by at least two external to the management team reviewers. For this reason, Task leaders responsible for their delivery will have to prepare a draft at least three weeks in advance of the deadline of delivery and report to the Coordinator. The PMT will then identify and appoint a reviewer and ensure the draft is read and feedback is provided to the Task leader before submission of the Deliverables. This mechanism will apply to those Deliverables that consist of a written document output (e.g. report, guideline document, article, etc). The deliverables, expected times for review and deadlines will be clearly set in the detailed work planning document and will be reviewed by two internal reviewers among the project partners; reviews and corrections will be communicated to the relevant WP and task leaders. Most important deliverables that will be published in the project collection in the Research Ideas and Outcomes (RIO) journal.
Task 12.6. Ethics management [Lead: Pensoft].
Pensoft will coordinate the management of ethical issues and compliance with GDPR of personal data together with the concerned partners. Ethics deliverables will ensure the collection and storage of personal data follows EU GDPR rules and research activities adhere to international guidelines for social research, as well as H2020 ethical standards (as per the EU Ethics Appraisal. An Ethical Statement will be submitted to the EC (D9.1) by M2 to ensure all activities are conducted in full compliance with fundamental ethical principles. These include the implementation of principles and protocols seeking, where necessary ethical approval from relevant ethics review boards (ERB), gaining informed consent (e.g. provision of participant information leaflets and informed consent forms) and the secure storage of personal data (e.g. anonymized, stored in a non-identifiable format, kept securely and shared for study purpose only in aggregated forms). Sensitive personal information, such as gender related issues, political opinion or religious convictions falls outside the scope of the BiCIKL project and will not be collected. The election of an Ombudsperson will guarantee that good ethics practices are upheld in the project. Gender and other equality issues will be monitored and regulated by a Equality and diversity champion appointed by the GA.
List of deliverables
- D12.1. Project Work Plan and Standard Operational Procedures (detailed description of governance structure & mechanisms) (M1)
- D12.2. Inception Report (detailed plans of the implementation developed during the Kick-off meeting) (M1)
- D12.3. General Assembly/Kick off meeting report (M2)
- D12.4. Second General Assembly report (M18)
- D12.5. Third General Assembly report (M34)
- D12.6. Internal communication platform (M4)
- D12.7. Risk management reviews I: risk register maintained and updates communicated to Executive Board and beneficiaries (M18)
- D12.8. Risk management reviews II: risk register maintained and updates communicated to Executive Board and beneficiaries (M36)
- D12.9. Data Management Plan (M6)
List of milestones
- MS45 Advisory Board Members appointed (M3)
- MS46 Kick off meeting (M1)
- MS47 Second Annual GA meeting held (M18)
- MS48 Third Annual GA meeting held (M34)
Management structure and procedures
General Assembly
The highest decision-making body of the project will be the General Assembly. Once a year, the General Assembly (GA) will bring together senior representatives from all beneficiaries Fig.
The GA will be open to the Advisory Board members who will receive special invitations for participation in interactive sessions with the project participants, thus being able to discuss various aspects of the project in terms of concept, methodology, strategic and technical issues and horizontal priority topics.
The General Assemblies will be the decision making body for:
- content, finances and intellectual property rights;
- evolution of the consortium;
- appointment of Executive Board Members (and if necessary the replacements for co-coordinator and WP lead- ers) and of members of additional project bodies (Advisory Board, Access Provision Panel);
- preparing and amending the Consortium Agreement.
Executive Board
Although the Coordinator is ultimately responsible for project leadership and implementation, it will be done in close collaboration with the Executive Board. The Executive Board of eleven members is formed by the Coordinator, and the ten work package leaders. Three of the WP leaders will also serve as Pillar leaders, with broader responsibility over and across their work packages, as follows: Pillar 1 - Networking Activities (Dimitris Koureas, Naturalis), Pillar 2 - Access activities (Jeremy Lanfear, ELIXIR Hub), Pillar 3 - Joint Research Activities (Quentin Groom, MBG). The role of the Pillar leaders includes in addition to coordination of task delivery within their work packages, the coordination of workflows between Pillars and across several WPs. The Coordinator and the Pillar leaders will thus maintain an overall management view over the entire project and will be in close contact. All WPs will appoint also co-leads at the start of the project to minimise risk and ensure smooth coordination within the consortium (Fig.
The Executive Board acts as the supervisory body for the execution of the Project and shall report to and be accountable to the General Assembly. The Executive Board, comprising the Project Coordinator, Pillar Leaders and Work Package Leaders, will meet via teleconference every month. This group will oversee the activities across all work packages, evaluate deliverables and other project outcomes and make the decisions affecting the day-to-day running of the project. The Executive Board will also review the attainment of Deliverables (Table
|
Deliverable number |
Deliverable title |
WP number |
Lead beneficiary |
Type |
Dissemination level |
Due date in months |
|
D1.1 |
User requirements analysis report |
WP1 |
LIFEWATCH |
Report |
Public |
12 |
|
D1.2 |
Recommendations for interoperability among infrastructures |
WP1 |
MeiseBG |
Report |
Public |
10 |
|
D1.3 |
Best practice manual for findability, re-use and accessibility of infrastructures |
WP1 |
NATURALIS |
Report |
Public |
18 |
|
D2.1 |
Relational network graph for the Biodiversity Knowledge Hub (BKH) |
WP2 |
GBIF |
Other |
Public |
15 |
|
D2.2 |
Stakeholder input into the network graph provided by Third Global Biodiversity Informatics Conference (GBIC3) |
WP2 |
GBIF |
Report |
Public |
15 |
|
D2.3 |
BiCKL training programme (MOOC) |
WP2 |
CETAF |
Demonstrator |
Public |
30 |
|
D3.1 |
Project logo, marketing pack and website design and development |
WP3 |
PENSOFT |
Report |
Public |
3 |
|
D3.2 |
Plan for dissemination and exploitation of project results |
WP3 |
CETAF |
Report |
Public |
7 |
|
D3.3 |
Outreach initiatives report |
WP3 |
CETAF |
Report |
Public |
36 |
|
D3.4 |
The BIodiversity Knowledge Hub (BKH) |
WP3 |
LIFEWATCH |
Demonstrator |
Public |
24 |
|
D4.1 |
Project call definition and execution plan, published via BiCIKL website |
WP4 |
ELIXIR/EMBL-EBI |
Websites, patents filing, etc. |
Public |
3 |
|
D4.2 |
Report on pilot call process |
WP4 |
ELIXIR/EMBL-EBI |
Report |
Public |
24 |
|
D4.3 |
Report on full call process |
WP4 |
ELIXIR/EMBL-EBI |
Report |
Public |
36 |
|
D4.4 |
Report on a long term sustainability plan for the BiCIKL community service |
WP4 |
ELIXIR/EMBL-EBI |
Report |
Public |
36 |
|
D5.1 |
World class virtual access to the infrastructures in support of Biodiversity and related subject matter research is provided |
WP5 |
ELIXIR/EMBL-EBI |
Demonstrator |
Public |
36 |
|
D5.2 |
Periodic assessment of VA by external board |
WP5 |
ELIXIR/EMBL-EBI |
Report |
Public |
36 |
|
D6.1 |
Enhanced, standalone and freely accessible installation of the ARPHA Writing Tool (AWT) |
WP6 |
PENSOFT |
Other |
Public |
24 |
|
D6.2 |
Automated workflow for discovery, semantic enhancement,mapping, extraction, FAIRfying, and dissemination of data liberated from literature |
WP6 |
Plazi |
Other |
Public |
36 |
|
D6.3 |
A workflow for bidirectional linking of data from literature to external resources |
WP6 |
Plazi |
Other |
Public |
18 |
|
D6.4 |
Applications for interoperable access to OpenBiodiv through semantically enhanced queries |
WP6 |
PENSOFT |
Other |
Public |
20 |
|
D7.1 |
Architecture Design for a pan-European PID system for Digital Specimens |
WP7 |
NATURALIS |
Demonstrator |
Public |
18 |
|
D7.2 |
Data standards and workflows for linking sequence data with specimens implemented and published |
WP7 |
FUB-BGBM |
Report |
Public |
30 |
|
D7.3 |
A repeatable informatics workflow that links people and places to stable identifiers of those entities, including a validation and preservation step |
WP7 |
MeiseBG |
Other |
Public |
30 |
|
D7.4 |
Digital Object Interface Protocol [DOIP] enabled Digital Object repository installation to store and provide digital specimen information |
WP7 |
NATURALIS |
Other |
Public |
36 |
|
D8.1 |
Availability via API of single, harmonised sequenced material source annotations in ENA |
WP8 |
ELIXIR/EMBL-EBI |
Other |
Public |
20 |
|
D8.2 |
Validation tool opensource software release |
WP8 |
ELIXIR/EMBL-EBI |
Other |
Public |
18 |
|
D8.3 |
Web interface for ELIXIR Contextual Data Clearinghouse |
WP8 |
UTARTU |
Other |
Public |
9 |
|
D8.4 |
First submission and access tools available for BIOM data |
WP8 |
ELIXIR/EMBL-EBI |
Other |
Public |
24 |
|
D9.1 |
POPD - Requirement No. 1 (Ethics) |
WP9 |
PENSOFT |
Ethics |
Confidential,only for membersof the consortium(including theCommissionServices) |
1 |
|
D10.1 |
New and updated taxonomic data products automatically visible in taxonomic framework |
WP10 |
GBIF |
Other |
Public |
30 |
|
D10.2 |
Workflow for bidirectional linking of taxonomic references to literature |
WP10 |
GBIF |
Other |
Public |
24 |
|
D10.3 |
Validation and curation workbench for taxonomic data |
WP10 |
sp2000 |
Other |
Public |
18 |
|
D10.4 |
Web visualisation of mappings between CoL and other checklists |
WP10 |
sp2000 |
Other |
Public |
12 |
|
D10.5 |
CoL services for direct encapsulated use within search and discovery services of BiCIKL partner infrastructures |
WP10 |
sp2000 |
Other |
Public |
27 |
|
D11.1 |
Evaluation benchmark: a database of questions associated with a set of relevant articles (e.g. PMIDs) to measure the progress of the WP towards an effective search service |
WP11 |
Plazi |
Other |
Public |
12 |
|
D11.2 |
Search and link association services: A RESTful API, which will input a link/ accession number and return a ranked list of neighbors links with a confidence score |
WP11 |
NATURALIS |
Demonstrator |
Public |
18 |
|
D11.3 |
Passage retrieval services: A RESTful API, which will input a set of keywords and return a set of articles/ passages ranked by relevance |
WP11 |
SIB |
Demonstrator |
Public |
18 |
|
D11.4 |
Factoid questionanswering: A RESTful API, which will input a factoid question in natural language and extract a short answer out of the articles/ passages returned by D10.3 |
WP11 |
SIB |
Demonstrator |
Public |
24 |
|
D11.5 |
Workbench implementation: a set of responsive Graphic User Interfaces developed in HTML 5.x/ JavaScript to operate the aforementioned services |
WP11 |
Plazi |
Demonstrator |
Public |
32 |
|
D12.1 |
Project Work Plan and Standard Operational Procedures |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
1 |
|
D12.2 |
Inception Report |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
1 |
|
D12.3 |
First General Assembly/Kick off meeting report |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
2 |
|
D12.4 |
Second General Assembly report |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
18 |
|
D12.5 |
Third General Assembly report |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
34 |
|
D12.6 |
Internal communication platform |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
4 |
|
D12.7 |
Risk management review I: risk register maintained and updates communicated to Executive Board and beneficiaries |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
18 |
|
D12.8 |
Risk management review II: risk register maintained and updates communicated to Executive Board and beneficiaries |
WP12 |
PENSOFT |
Report |
Confidential,only for membersof the consortium(including theCommissionServices) |
36 |
|
D12.9 |
Data Management Plan |
WP12 |
PENSOFT |
ORDP: Open Research Data Pilot |
Public |
6 |
|
Milestone number |
Milestone title |
Related work package |
Lead beneficiary |
Means of verification |
Due date (in month) |
|
MS1 |
The first BiCIKL data hackathon held with partners and selected invitees |
WP1 |
Lifewatch |
Meeting attendance list |
4 |
|
MS2 |
Third Global Biodiversity Informatics Conference (GBIC3) |
WP2 |
GBIF |
Meeting and presentation of draft network graph |
15 |
|
MS3 |
Network graph database |
WP2 |
GBIF |
Presentation and online dashboard |
15 |
|
MS4 |
Workshop for training needs identification |
WP2 |
GETAF |
Report on the Workshop outcomes |
20 |
|
MS5 |
Training Modules on data liberation or publishing tools pilot activity |
WP2 |
CETAF |
Modules uploaded in the BiCIKL Tool Platform |
29 |
|
MS6 |
Best Practices and recommendations (from T3.3) translated into training material |
WP2 |
CETAF |
Report uploaded in the BiCIKL Tool Platform |
29 |
|
MS7 |
Communication strategy |
WP3 |
Pensoft |
Report |
6 |
|
MS8 |
Biodiversity Knowledge Hub prototype |
WP3 |
Lifewatch |
Website |
12 |
|
MS9 |
Dissemination plan revision |
WP3 |
CETAF |
Report |
24 |
|
MS10 |
Workshop with RIs |
WP3 |
CETAF |
Meeting |
24 |
|
MS11 |
Workshop with publishers |
WP3 |
CETAF |
Meeting |
28 |
|
MS12 |
Biodiversity Knowledge Hub Best Practices communicated to T2.3 |
WP3 |
Lifewatch |
Report |
29 |
|
MS13 |
Outreach activities report I |
WP3 |
CETAF |
Report |
12 |
|
MS14 |
Outreach activities report II |
WP3 |
CETAF |
Report |
24 |
|
MS15 |
Project call process definition phase completed |
WP4 |
ELIXIR/EMBL-EBI |
Report |
3 |
|
MS16 |
Pilot call projects process opens |
WP4 |
ELIXIR/EMBL-EBI |
Call for pilot project proposals is made public via the BiCIKL web site and other channels |
12 |
|
MS17 |
Full call projects process opens |
WP4 |
ELIXIR/EMBL-EBI |
Call for full project proposals is made public via the BiCIKL web site and other channels |
24 |
|
MS18 |
Initial teleconference between infrastructures and follow-on regular TCs will have been established and set up |
WP5 |
ELIXIR/EMBL-EBI |
Meeting minutes |
3 |
|
MS19 |
External board review received I |
WP5 |
ELIXIR/EMBL-EBI |
Report |
12 |
|
MS20 |
External board review received II |
WP5 |
ELIXIR/EMBL-EBI |
Report |
24 |
|
MS21 |
External board review received III |
WP5 |
ELIXIR/EMBL-EBI |
Report |
36 |
|
MS22 |
Specification of the independent authoring and editing environment |
WP6 |
Pensoft |
Report |
6 |
|
MS23 |
Initial scoping and assessment of optimal degree of automation for processing workflow |
WP6 |
Plazi |
Report |
9 |
|
MS24 |
Prototype implementation of proposed advanced automation tools |
WP6 |
Plazi |
Product |
21 |
|
MS25 |
Beta-implementation and deployment of automated data extraction and preparation service |
WP6 |
Plazi |
Product |
30 |
|
MS26 |
Scoping and specifications for workbench |
WP6 |
Plazi |
Report |
12 |
|
MS27 |
App specifications |
WP6 |
Pensoft |
Report |
10 |
|
MS28 |
Architecture design ready for evaluation by potential end users; design and specification report. |
WP7 |
Naturalis |
Report |
9 |
|
MS29 |
PID system development roadmap and technical provisioning for test system (beta/pre-production) |
WP7 |
Naturalis |
Roadmap, beta PID system |
14 |
|
MS30 |
Draft recommendation for specimen links in sequence data |
WP7 |
FUB-BGBM |
Report |
24 |
|
MS31 |
Full data and API specification for specimen links in sequence data |
WP7 |
FUB-BGBM |
Report |
27 |
|
MS32 |
The design and prototype of a workflow integrating Wikidata into validation and linking |
WP7 |
MeiseBG |
Report |
25 |
|
MS33 |
Documentation on how to interface with FAIR Digital Object via Digital Object Interface Protocol (DOIP) |
WP7 |
Naturalis |
Documentation |
27 |
|
MS34 |
Report on profile of sequenced material source annotations |
WP8 |
EMBL |
Report |
6 |
|
MS35 |
Submission for publication of “application note” relating to sequenced material source validation tool |
WP8 |
ELIXIR/EMBL-EBI |
Manuscript |
14 |
|
MS36 |
First improved sequenced material source annotations routed through the ELIXIR Contextual Data Clearinghouse |
WP8 |
UTARTU |
News item |
6 |
|
MS37 |
Specification for curation workbench |
WP10 |
GBIF |
Report |
12 |
|
MS38 |
Implementation of workbench released for use by taxonomic users |
WP10 |
GBIF |
Product |
21 |
|
MS39 |
Specification for encapsulated services |
WP10 |
Sp2000 |
Report |
18 |
|
MS40 |
Integration of encapsulated service into first partner infrastructure |
WP10 |
Sp2000 |
Product |
27 |
|
MS41 |
Implementation of mapping visualisation released |
WP10 |
Sp2000 |
Product |
27 |
|
MS42 |
Gathering of a limited set of biodiversity-related questions |
WP11 |
Plazi |
CSV database |
6 |
|
MS43 |
Mockups to support the design of the link association services |
WP11 |
Naturalis |
Presentation |
15 |
|
MS44 |
Mockups to support the design of the workbench |
WP11 |
Plazi |
Presentation |
28 |
|
MS45 |
WP12 |
Pensoft |
Letters of intent |
3 |
|
|
MS46 |
WP12 |
Pensoft |
Meeting report at M2 |
1 |
|
|
MS47 |
Second Annual GA meeting held |
WP12 |
Pensoft |
Meeting report |
18 |
|
MS48 |
Third Annual GA meeting held |
WP12 |
Pensoft |
Meeting report |
34 |
Another specific function of the Executive Board in relation to Trans-national Access applications is to take decisions on the joint provision of access to the participating RIs. This latter function will be undertaken in implementation of the decisions of the Access Provision Panel (see Trans-national Access).
Budget and time tolerances will be controlled at each management level. Task Leaders will alert WP Leaders if there is a risk their work will deviate from budget, scope or timing. If this exceeds the agreed tolerances, WP Leaders will inform the Project Manager who will advise and in turn escalate to Pillar Leaders coordinators if required. Pillar Leaders will report any risks, issues or deviations at the monthly Executive Board meetings, and matters will be escalated to the General Assembly if required. Milestones (Table
- coordinate and facilitate interactions and integration between Pillars, WPs and Tasks;
- collect information on the progress of the project, assess the compliance of the project with its objectives and, if necessary, propose modifications to the GA;
- support the Coordinator in preparing meetings with the Funding Authority and in preparing related data and deliverables;
- prepare the content and timing of press releases and joint publications by the consortium or proposed by the Funding Authority;
- any other activities specified in the Consortium Agreement or required by the GA.
To follow the progress of the project by the Executive Board, General Assembly and the Contracting Authority a list of milestones has been developed and presented in List of milestones (Table
Project Coordination
The Coordinator of BiCIKL is Pensoft Publishers Ltd. (Pensoft) - the legal entity acting as the intermediary between the Parties and the Funding Authority and responsible for the overall performance and coordination of the project. The Coordinator shall, in addition to its responsibilities as a Party, perform the tasks assigned to it as described in the Grant Agreement and the Consortium Agreement. The Coordinator Pensoft is represented by its Managing Director Prof. Dr. Lyubomir Penev, acting as Project Coordinator.
The Project Coordinator will be supported by the Management Support Team, employed by Pensoft, who will be responsible for implementing the daily tasks of project management, administration, finance and communication. The Management Support Team assists the Project Coordinator and the Executive Board in the day to day running of the project. Furthermore, the PMT will assist the Coordinator in:
- contacting and engaging with the Consortium members;
- providing administrative and legal management to ensure compliance with the Grant Agreement;
- maintaining and coordinating financial records including cost statement submissions by all partners according to the Grant Agreement;
- coordinating periodic reporting and EC review.
The team comprises of three officers responsi- ble for Project Management, Administration & Finance and Communication & Dissemination:
- Project Manager: The Project Manager is the first point of contact for all work package related matters, and will be responsible for organising project management and other operational meetings and events as well as for receiving, registering, reviewing and distributing project deliverables and milestones.
- Project Administration & Financial Manager: The effective administration and financial management of BiCIKL will be the responsibility of Project Administrative and Finance Manager. His/Her duties will cover the management of administrative and financial tasks by coordinating the establishment of the financial reporting and audit procedures across the BiCIKL consortium, in line with EU requirements, Grant Agreement and Consortium Agreement.
- Project Communication & Dissemination Manager: The Project Communication and Dissemination plans will be developed, overseen and implemented by the Project Communication & Dissemination Manager, responsible for the implementation of the project communication strategy which includes internal and external communication objectives and actions.
The Management Support Team will work in close coordination and collaboration, ensured by weekly team meetings and a monthly Project Management Review meeting.
The Project Coordinator, with the assistance of the Management Support Team, is responsible for:
- monitoring compliance by the Parties with their obligations and execution of the GA decisions;
- representing the intermediary for communication between the EC and the consortium;
- collecting, reviewing and submitting reports, other deliverables (including financial statements and related certifications) and specific requested documents to the EC;
- administering the financial contribution of the EC and financial tasks as described in the Consortium Agreement;
- implementing efficient project management, provide management in all administrative, legal, financial, and scientific matters, and ensuring the day-to-day project management;
- chairing and organising the GA and SC meetings and drafting the minutes;
- working closely together with the NA-03 (WP3) lead and partners to ensure an efficient dissemination of information and outreach beyond the Consortium towards the larger scientific community, international networks, stakeholders, and SMEs and implementing and updating the BiCIKL project website.
An overall agile management approach will be developed, so that regular reviews of progress can feed into the project implementation plans and, if necessary, adjustments in the plans be made.
At the first GA meeting an Ombudsperson will be elected among the project partners to guarantee that good ethics practices are upheld in the project.
To ensure a better coordination and management of critically important issues across the whole project, three specific roles are introduced and their occupation is proposed in the project description:
- Equality and diversity champion (Dr Ana Casino, CETAF);
- Innovation champion (Dr Donat Agosti, Plazi);
- Open science champion (Prof. Lyubomir Penev, Pensoft).
Further, to ensure the smooth implementation and to avoid delays and gaps in the delivery of results, the following risks were identified and classified according to their level of likelihood and together with the proposed measures for mitigation.
Additional structures
Advisory Board (AB): The AB will be composed of minimum five members, all internationally recognized scientists in the field listed below. The AB role is crucial for the governance of the Consortium. It will report directly to the project’s General Assembly and on an ad-hoc basis to the Executive Board in case their advice is required to tackle specific topics. The areas of primary interest to BiCIKL and prospective members of the Advisory Board (to be invited after the start of the project) are:
- Biodiversity informatics standards: James Macklin, Agriculture and Agri-Food Canada; Steve Baskauf, Vanderbilt University, USA
- Text and data mining, web annotations: Peter Cornwell, Data Futures, University of Westminster, UK and École normale supérieure lettres et sciences humaines Lyon, France
- Biodiversity genomics (especially semantics): Dr. Pier Luigi Buttigieg, Max Planck Institute for Marine Microbiology, Bremen, Germany
- FAIR data management and library science: Dr. Alex Hardisty, University of Cardiff. Mark D Wilkinson; Flo- ra D’Anna (+VIB-UGent Center for Plant Systems Biology); Heike Neuroth, University of Applied Sciences Potsdam, Potsdam, Germany
- Taxonomy data flows: Dr. Conrad Schoch, NCBI Taxonomy, NIH-NCBI, Bethesda
- Reproducible science: Patricia Herterich, University of Edinburgh, UK; Dr. Sabina Leonelli, Exeter Centre for the Study of the Life Sciences (Egenis), UK
- Agricultural open science: Odile Hologne, Institut national de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), Paris, France
Access Provision Panel (APP): The APP is responsible for the selection of researchers or research teams through an independent peer-review evaluation of their research projects. The Panel is composed of seven scientists appointed by the TA and VP WP Leader (ELIXIR Hub) and the research infrastructures providing Trans-national and Virtual Access. The selection of researchers or research teams shall be carried out through an independent peer-review evaluation of their research projects. Each project will be evaluated by 3 independent external reviewers, who will evaluate the proposals against pre-defined selection criteria. The reviews will be used to inform the internal BiCIKL panel who will make the final project selection based on both scientific and logistical considerations. This is to allow for the fact that certain projects may score scientifically very high, but nevertheless would be difficult to implement within the resource window of the BiCIKL project.
Operational procedures
The organisational structure of BiCIKL alongside all management structures and their decision-making procedures are described in the subsections above. The operational procedures of BiCIKL will be further clarified and officially established in section 6.2 of the Consortium Agreement, which the project will sign upon eventual successful evaluation. A detailed SOP document will be adopted by the first GA.
Consortium as a whole
Expertise and complementarity
The BiCIKL consortium consists of 14 partners from 9 countries. The consortium represents different kinds of organisations, namely large international networks of research-supporting organisations (GBIF, LIFEWATCH, CETAF, CERN, ELIXIR Hub, EMBL, TDWG), universities, institutes, museums, botanical gardens (Naturalis, SIB, MBG, FUB-BGBM, UTARTU), and SMEs (Pensoft, Plazi). All these organisations are complementary to each other in that they place emphasis on different aspects of establishing open science research practices in the domain of biodiversity and related areas of molecular biology. These subject areas include:
- zoology, botany (incl. algae) and mycology, molecular systematics, evolution, systems biology, genomics, proteomics, transcriptomics and metabarcoding;
- ecology, conservation, population genetics, biological invasions;
- structural and evolutionary bioinformatics, modelling, imaging, data standardization, computing management and data management, software development and web design;
- large-scale digitisation of legacy literature, text and data mining, academic open access book and journal publishing, indexing, project dissemination and science communication;
-
citizen science in biodiversity;
-
collection management.
The consortium will work in a truly interdisciplinary and integrative manner. It is therefore well suited to successfully complete the project work in terms of variety and depth of knowledge and experience. The partners possess the necessary key qualifications, including project management, education/training, stakeholder engagement, dissemination and science-policy interface at national, European and global levels.
As an essential element of the project, all work packages are designed to have clear links and specified hand-over points between each other and with the responsible beneficiaries (Fig.
Through the involvement of global or supranational networks of research institutions such as GBIF, LIFEWATCH, CETAF and TDWG, BiCIKL will guarantee high quality and diverse services and expertise to its end users. Moreover, most of the key partners have sound experience in coordinating RIs at European (DiSSCo, Elixir Data Platform, SYNTHESYS+) and national level (NATARC: Estonian research infrastructure roadmap project “Natural history archives and information network”4, SVIP-O: Swiss Variant Interpretation Platform5; TrIAS (Belgian Science Policy Office) Tracking Invasive Alien Species6). The experience acquired through these projects will be invaluable for ensuring successful implementation, as well as for identifying the most efficient ways to achieve the objectives of this proposal. BiCIKL represents a broad range of disciplines and engages several high-level specialists providing a comprehensive matrix of complementary skills and experience. This allows effective trans-disciplinary tasks to be conducted. The complementary skills also ensure the completion of the project objectives with the help of potential backup plans.
The key competences of the consortium partners as a whole cover all major areas and include taxonomy and systematics, genetics, bioinformatics and more applied and systems-oriented areas such as modelling, data standardization, software development, literature digitization, open-access book and journal publishing. These are supplemented by strong organizational, communication and training skills. The consortium expertise is widely recognised by the scientific community and will enable several innovative technologies and services to be developed in WP JRA-01 to JRA-05.
BiCIKL will have an important role in training a new generation of taxonomists, molecular biologists, bioinformaticians and collection managers. One of the great ambitions of the project is to develop a modern, multi-functional, communication and publishing platform, the Biodiversity Knowledge Hub (BKH) and for the first time, a federated search tool, the Fair Data Place (FDP). This tool will bring the state of knowledge a big step forward in terms of data sharing and open data access, which will be highly beneficial for the whole emerging community.
BiCIKL brings together world leading partners in the field of information systems for large scale data and metadata management (GBIF, ELIXIR Hub, EMBL-EBI, Sp2000). A unique feature of BiCIKL within the scientific landscape is its perennial emphasis on collection management activities and collaborations between natural scientists (CETAF, Naturalis, FUB-BGBM, MBG) in biodiversity and ecosystem management (LIFEWATCH) and training capacity (CETAF). The BiCIKL genomics research will go beyond by bringing together leading experts in genomics, proteomics, transcriptomics and metabarcoding (EMBL-EBI, ELIXIR Hub, SIB, UTARTU). Consortium expertise will be strengthened by the involvement of renowned open access publishers (Pensoft) and literature digitalization specialists (Plazi).
Industrial and commercial involvement
Two BiCIKL beneficiaries are SMEs (Pensoft and Plazi). Pensoft will act as a project coordinator, communication and dissemination expert. Pensoft brings their proven expertise in developing innovative publishing tools and workflows, including data auditing and publishing. Apart from that, the SME will be actively involved in various technology development activities related to Linked Open Data and next-generation publishing tools. Creating a uniquely valuable LOD-based knowledge graph for biodiversity science will increase the company’s capacity and provide new business opportunities, which will place it among the leaders in the academic publishing market.
Plazi will lead the WP6 (JRA-01) and will be involved in networking activities. The company will provide trans-national access to the TreatmentBank data and virtual access to the Biodiversity Literature Repository. By developing a highly automatic pipeline for large scale literature digitization, text and data mining, and management of liberated data, Plazi will strengthen its unique leading position in the domain.
International involvement of the participants
Most beneficiaries have active links to international projects and RIs, and thus ensure that BiCIKL is in line with and contributes to European and global initiatives and processes. Below, a categorization is provided:
- Global networks: GBIF, TDWG, Sp2000
- European networks: CERN, CETAF, LIFEWATCH, EMBL-EBI, ELIXIR Hub
- National Public bodies: UTARTU, SIB, MBG, FUB-BGBM, CETAF-MNHN
- SMEs, operating on a global level: Pensoft, Plazi
Members of the consortium
Partner 1: Pensoft Publishers (Coordination)
Description of the legal entity
Pensoft Publishers is a SME specializing in academic, open access book and journal publishing, software development and web design, project dissemination and science communication.
The company’s project department consists of a motivated team of active scientists, project managers and science communicators. Among the services for projects offered by Pensoft are: 1) Development of project logo and brand identity; 2) website design, setup and maintenance; 3) setup of project management tools: internal communication platform, storage and mailing modules; 4) design, production and distribution of marketing collateral: flyers, posters, stickers, videos, other branded products; 5) Organization of events, workshops, summer schools; 6) Consultancy and development of communication strategies, plans for dissemination and exploitation and data management plans; 7) Press release writing and dissemination and liaison with journalists; 8) Social media setup and management; 9) Production and distribution of final results packages (policy briefs, factsheets, infographics) and booklets; 10) Design and development of interactive final project online information resources and tools.
Throughout the last 20 years, Pensoft has been actively involved in managing, planning and carrying out dissemination and communication activities for several EU projects. Among these are STEP, ALARM, MOTIVE, SCALES, BIOFORUM, MACIS, COCONUT, MACMAN, EURO-LIMPAKS, RUBICODE, pro-iBiosphere, BESAFE, EU BON, ESMERALDA, IMPRESSIONS, STACCATO, Super-B, BIG4, NanoFASE and AGINFRA PLUS. Currently, Pensoft’s staff is participating as a WP Leader in the Horizon 2020 projects CLAIM, PoshBee, RENATURE, HOMED, Path2Integrity, eLTER PPP, eLTER PLUS and SHOWCASE. Pensoft is also involved as a dissemination partner in the H2020 projects IGNITE, BESTMAP, B-GOOD and MAIA.
Since its foundation in 1994, Pensoft has published more than 1000 books and e-books. In 2014 the company launched the novel ARPHA Publishing Platform, which serves Open Access academic Journals, Books, and Conference Materials. Pensoft is well known among academics worldwide with its technologically advanced peer-reviewed Open Access journals, such as ZooKeys, PhytoKeys, MycoKeys, Nature Conservation, NeoBiota, Comparative Cytogenetics, Biodiversity Data Journal (BDJ). The company is actively developing new tools, workflows and methods for text- and data publishing, dissemination of scientific information and technologies for semantic enrichment of articles’ content. Pensoft is actively looking to expand the subject scope of its publishing towards open science publishing practices with the launch of Research Ideas and Outcomes (RIO) - an open science journal that publishes all research ideas & outcomes that constitute the research cycle, including: project proposals, data, methods, workflows, software, project reports and research articles.
Being and open access publisher, Pensoft can also consult in best practices in open access and open data publishing, while offering expertise and know-how in dissemination of scientific results through special issues in peer-reviewed journals, books and article collections (example: Scaling in Ecology and Biodiversity Conservation).
Main tasks of the entity in the project
In BiCIKL Pensoft will coordinate the project (WP12); contribute heavily to WP6 where new-generation semantic publishing tools and open science workflows will be developed; and will also be responsible for development of the project image, website, production of various outreach material (brochures, posters, leaflets) and contribute to the active dissemination of the project results (WP3).
Key persons assigned to the project
Prof. Dr. Lyubomir Penev (Male) is a Professor of Ecology at the Bulgarian Academy of Sciences, Sofia and Managing Director of Pensoft Publishers. His main interests over the past 30 years have been the methods of biodiversity study, development of software for biodiversity research and environmental assessment, biogeography, urban ecology, data publishing and management. He has published more than 130 papers and was co-author or (co-)editor of 8 books. He is involved as a work package leader in several FP7 and Horizon 2020 projects dealing with mobilization and integration of biodiversity data and development of e-infrastructure. Web of Science: 989 citations, H-Index 16; SCOPUS: 1311 citations, H-Index: 19; Google Scholar: 3687 citations; H-Index 30.
Prof. Dr. Pavel Stoev (Male) is a Professor at the National Museum of Natural History, Bulgaria and Head of the Projects department at Pensoft Publishers. His research interests include systematics and biogeography of cave and soil-dwelling arthropods, distribution of invasive species, bioinformatics, and data management. His research combined taxonomic and ecological knowledge with bioinformatics to develop innovative publishing models and workflows with Pensoft’s open access journals. He has published more than 120 papers and 3 monographs. He has been a project leader of several conservation and scientific projects. Web of Science: 552 citations, H-Index 15; SCOPUS: 507 citations, H-Index: 11; Google Scholar: 1921 citations; H-Index 22.
Teodor Georgiev (Male) has more than 15 years of experience in designing and desktop publishing of nearly 600 books. He is highly experienced in several software packages, such as Adobe InDesign, Photoshop, Illustrator, MS Office, and others, as well as in projecting and designing web-based platforms, mark up of legacy literature, online journal publishing and dissemination. T. Georgiev has published ca. 20 papers in bioinformatics and semantic publishing.
Iliyana Demirova (Female) is a marketing and public relations specialist currently acting as the Head of Press Office at Pensoft. She has specialised and gained extensive experience in science communication, including press release writing and distribution, liaising with international media, and preparation of promotional materials. She is involved in the dissemination and communication of a number of EU funded projects, where she is participating actively in the preparation and implementation of long-term, multi-channel Communication and Dissemination Strategies.
Boris Barov (Male) has a MSc in Ecology and 20 years of professional experience in biodiversity, nature conservation and business sustainability at international level. As programme manager in BirdLife International, Boris successfully coordinated contracts with the European Commission and international conventions. He has led the development, evaluation and adoption of over 30 international species action plans and has authored methodologies, guidelines and policy papers for the EC. He is an experienced team leader of non-profit, academic and industry partners credited for his diplomacy, advocacy and stakeholder involvement skills. He has been regularly invited to evaluate Horizon 2020 and other EU funded projects. B. Barov has joined Pensoft in 2020 as Project Manager.
Margarita Grudova (Female) has more than 15 years of experience in coordination and management of projects in the field of environmental protection, funded under different international programmes. Before joining the Pensoft’s Projects department she worked as a Chief Expert at the Executive Environment Agency with the Bulgarian Ministry of Environment and Water. She has specialised in project financial management and control as well as administrative and financial reporting for FP7 and H2020 funded projects.
Relevant publicatins
- Penev L, Dimitrova M, Senderov V, Zhelezov, G, Georgiev T, Stoev P, Simov K (2019) OpenBiodiv: A Knowledge Graph for Literature-Extracted Linked Open Data in Biodiversity Science. Publications, 7(2) 38. https://doi.org/10.3390/publications7020038
- Burkhard B, et al. (2018) Mapping and assessing ecosystem services in the EU - Lessons learned from the ESMERALDA approach of integration. One Ecosystem 3: e29153. https://doi.org/10.3897/oneeco.3.e29153
- Penev L, Mietchen D, Chavan V, Hagedorn G, Smith V, Shotton D, Ó Tuama É, Senderov V, Georgiev T, Stoev P, Groom Q, Remsen D, Edmunds S (2017) Strategies and guidelines for scholarly publishing of biodiversity data. Research Ideas and Outcomes 3: e12431. https://doi.org/10.3897/rio.3.e12431
- Hoffmann A, Penner J, Vohland K, Cramer W, Doubleday R, Henle K, Kõljalg U, Kühn I, Kunin WE, Negro JJ, Penev L, Rodríguez C, Saarenmaa H, Schmeller DS, Stoev P, Sutherland WJ, Tuama1 EO, Wetzel F, Häuser CL (2014) Improved access to integrated biodiversity data for science, practice, and policy - the European Biodiversity Observation Network (EU BON). Nature Conservation 6: 49–65. https://doi.org/10.3897/natureconservation.6.6498
- Chavan V, Penev L (2011) The data paper: a mechanism to incentivize data publishing in biodiversity science. BMC Bioinformatics, 12: S2. https://doi.org/10.1186/1471-2105-12-S15-S2
Relevant previous projects
- pro-iBiosphere (FP7 grant 312848): Coordination and policy development in preparation for a European Open Biodiversity Knowledge Management System, addressing Acquisition, Curation, Synthesis, Interoperability and Dissemination, http://www.pro-ibiosphere.eu, timing: 2012-2014.
- EU BON (FP7 grant 308454): EU BON: Building the European Biodiversity Observation Network, http://eubon.eu, timing: 2012-2017.
- IMPRESSIONS (FP7 grant 603416): Impacts and risks from higher-end scenarios: Strategies for innovative solutions, http://www.impressions-project.eu, timing: 2013-2018.
- AGINFRA PLUS (H2020 grant 731001): Accelerating user-driven e-infrastructure innovation in Food Agriculture, http://plus.aginfra.eu, timing: 2017-2019.
- eLTER PLUS (H2020 grant 871128): European long-term ecosystem, critical zone and socio- ecological systems research infrastructure PLUS, timing: 2020-2025.
Partner 2: Naturalis Biodiversity Center
Description of the legal entity
Naturalis Biodiversity Center chairs the coordination team leading the development of the DiSSCo RI. It is one of the largest natural history museums in the world and the leading institute in the Netherlands for academic research and education on biodiversity and taxonomy. Naturalis maintains the national natural history collection of the Netherlands, a scientific collection of 42 million zoological, botanical and geological specimens. In a five-year program that ended in 2015, a cross-section of 8 million specimens from all collection types has been digitised on specimen object level with the remaining 34 million on storage unit level, using a large-scale industrial approach.
The collection is the focal point for the 120 employed researchers and a large number of research associates, research fellows and PhD students, who work on various fundamental and applied biodiversity research topics, programmes, such as dynamic biodiversity and societal impact, evolution of species interdependencies and character evolution, as well as in systematic and applied research, yielding an annual output of over two hundred scientific publications. The generated knowledge is translated for the general public into museum exhibits, teaching programmes and digital resources. Through the ICT department, Naturalis has built a strong presence within the biodiversity informatics community, producing high- standard software for managing and applying heterogeneous biodiversity data and information (such as Linnaeus NG and BioPortal), as well as researching and developing innovative digital information products. The collection is a source of study for the over 700 visiting researchers yearly from all over the world and the open data on platforms such as GBIF is available for scientists and conservationists worldwide. Naturalis
also hosts and chairs the secretariat of Species 2000 that together with the Integrated Taxonomic Information System (ITIS) forms the Catalogue of Life Partnership.
Main tasks of the entity in the project
Naturalis (Dimitris Koureas) is coordinator of the Networking Pillar of BiCIKL, Naturalis is leading the work under JRA-02 (WP7), as well as, Tasks 7.1 and 7.4 in JRA-02 and 10.2 in JRA-05. Involvement in Tasks of NA 2, 3 and 4 of the Networking Pillar. In the project Naturalis is additionally spearheading the DiSSCo Virtual Access activities.
Key persons assigned to the project
Dr Dimitris Koureas (Male) is head of the department for the development of international biodiversity Research Infrastructures at Naturalis and the coordinator of the DiSSCo Research Infrastructure.
He holds a PhD in plant systematics with post-doctoral experience acquired in biodiversity informatics/e-taxonomy. A DAAD scholar, awarded member of the Hellenic Botanical Society and elected member of the Linnean Society. Dr Koureas has previously led the development and implementation of several European Commission co-funded research and infrastructure projects (incl. ICEDIG, SYNTHESYS+ and DiSSCo Prepare & Mobilise COST Action). He serves on the Technical Advisory Board of the Research Data Alliance (RDA) and is a member of the Coordination Group for FAIR Digital Objects (FDO). Invited lecturer to MSc programmes (incl. University of Oxford and Reading University). He served as President of the international organisation for Biodiversity Information Standards (TDWG).
Dr Laurens Hogeweg (Male) is leading the automatic species identification project at Naturalis and is at COSMONiO Imaging BV responsible for the research and development of NOUS, an interactive deep learning-based platform for rapid annotation and validation of datasets. Within COSMONiO Laurens specializes in active machine learning, i.e. methods that aim to reduce the human annotation effort by performing a smart selection of data. Before joining COSMONiO and Naturalis he obtained a PhD in medical image analysis using machine learning at Radboud University. Laurens has ample experience in software development (25 years) and more than 10 years specifically in machine-learning-based (image) processing.
Dr Sharif Islam (Male) (B.Sc Math and Computer Science, University of Illinois 2003, PhD Sociology, University of Illinois 2016) has more than ten years of experience working with large scale research computing and data infrastructures in the USA and Europe. He is currently the Data Architect for DiSSCo, member of the DiSSCo technical team and leading the design of the European Loans and Visits system (ELViS) in the EU funded SYNTHESYS+ project. He also worked as a technical lead and system architect for research data management services at SURFsara (Dutch National Supercomputing Center). Prior to that, as the Lead System Engineer for the Blue Waters supercomputer (National Center For Supercomputing Applications in Urbana, Illinois, USA), he was responsible for maintaining system-wide resources and the whole software stack. Sharif is a member of the international technical implementation group on Fair Digital Objects (FDO) led by Rob Quick (Indiana University) and Luiz Bonino (GoFAIR).
Relevant publications
- Hogeweg L (2018) Reducing the taxonomist’s burden through AI. EuropeanaTech conference, Abstract Booklet, Rotterdam (the Netherlands), May 2018: 18. https://pro.europeana.eu/files/Europeana_Professional/Event_documentation/Events/EuropeanaTech_201 8/Speaker_PDF/Laurens-Hogeweg.pdf
- Hogeweg L (2018) Reducing the taxonomist’s burden through AI. EuropeanaTech conference, Abstract Booklet, Rotterdam (the Netherlands), May 2018: 18. https://pro.europeana.eu/files/Europeana_Professional/Event_documentation/Events/EuropeanaTech_201 8/Speaker_PDF/Laurens-Hogeweg.pdf
- Koureas D, Hardisty A, Vos R et al. (2016) Unifying European Biodiversity Informatics (BioUnify). Research Ideas and Outcomes 2: e7787. https://doi.org/10.3897/rio.2.e7787
- Addink W, Koureas D, Casino A. (2018) DiSSCo: The physical and data infrastructure for Europe's Natural Science Collections. 20th EGU General Assembly, EGU2018, Proceedings from the conference held 4-13 April, 2018 in Vienna, Austria, p.16356
- González-Aranda JM, Koureas D, Addink W, Hirsch T (2019) Facing e-Biodiversity Challenges Together: GBIO framework-based synergies between DiSSCo and LifeWatch ERIC. Biodiversity Information Science and Standards 3: e38554. https://doi.org/10.3897/biss.3.38554
Relevant previous projects
- MOBILISE COST Action CA17106 (2018-2022) - Mobilising Data, Policies and Experts in Scientific Collections. Naturalis is chairing the Action that will foster a cooperative network in Europe to support excellent research activities and facilitate knowledge and technology transfer around natural sciencecollections.
- Catalogue of Life Plus (2017-2019). Together with the Global Biodiversity Information Facility Secretariat and Species 2000 / Catalogue of Life, Naturalis is coordinating the Catalogue of Life Plus initiative to establish and maintain a global infrastructure for names and taxonomy in partnership with the Barcode of Life Data system, Biodiversity Heritage Library, and the Encyclopedia of Life.
- SYNTHESYS 1-3 (2004-2017) and SYNTHESYS Plus (2019-2022). Key partner in previous SYNTHESYS projects, providing Transnational Access and also participating in JRA and NA activities and leading a Joint Research Activity work package in the current project.
- DIOPSIS (Digital Identification of Photographically Sampled Insect Species) project which installed 100 smart cameras to monitor insects.
- Building the Databases of Life. Project funded by the Dutch Organisation for Scientific Research (NWO) aimed at providing large volumes of heterogeneous biodiversity data, among other methods through large scale natural history collections digitisation (2005-2013).
Partner 3: Plazi GmbH
Description of the legal entity
Plazi GmbH is a Swiss based SME providing services supporting and promoting the development of persistent and openly accessible digital taxonomic literature. Plazi GmbH was founded in 2012 as service provider of and completely owned by not-for-profit Plazi Verein, itself funded in 2008 as a spin off from a US National Science Foundation/German Science Foundation (DFG) binational research award to investigate the extraction of data from legacy taxonomic literature.
Plazi has lead the development of data sharing policies in the EU FP7 funded projects pro-iBiosphere and EU BON, has been involved in Globis-B related legal issues, has been task leader in the H2020 funded ICEDIG project, helped draft the RDA/CODATA legal interoperability principles and guidelines and, published in copyright issues related to biodiversity data and workflows.
Since 2004, Plazi has been developing and implementing open source tools and services to liberate, enhance, FAIRify , and disseminate data from taxonomic publications (The TreatmenBank data preparation services). Plazi has been leading the development of TaxPub, a taxonomic specific extension of the Journal Article Tag Suite (JATS) in collaboration with the US National Center of Biotechnology Information, and currently used by two publishers (Pensoft and EJT) for 30 journals and allowing automatic reuse of the data published therein.
Plazi’s TreatmentBank data preparation tools allows you to liberate, discover, and enhance automatically data in scholarly articles. Its tool to create templates describing the structure of scholarly articles (PDFs) allows automation of the service to process articles (2019: total production 5542 articles from 178 journals of which 26 are processed fully automatically, 65K taxonomic treatments, 30K figures). A pilot tool to quality control the output allows to monitor and curate the output.
The Biodiversity Literature Repository (BLR) developed together with Pensoft as a community within the Zenodo repository at CERN is providing access to the liberated data, including rich metadata based on standards developed in TDWG, mint digital object identifiers (DOI) and provide long term storage (270K deposits: 220K images, 50K publications) (Feb. 2020). Small taxonomic publishers are supported to mint DOIs for their articles to be published.
Within the recent OpenAIRE funded project (Advanced Canonical Person Resolution Service), a service will provide a canonical name for person names.
All the data produced is immediately reused by the Global Biodiversity Information Facility (GBIF) and is responsible to cut down the time from the publication of new species from months or years to hours after the production of the journals. Over 50K taxononomic names for new species are only supplied by Plazi. The US NCBI is furnished daily with a list of new taxonomic entities added to BLR.
A commercial service is offered for publishers to make their data contained in the PDFs FAIR.
Plazi is currently supported by a EUR1.1M award from the Arcadia Fund (2018-21) to liberate a large enough number of data (>50% of the annually newly discovered species) to convince the taxonomic community to switch to open access publishing.
Plazi has its main office in Switzerland, currently including 13 highly motivated employees or contractors).
Main tasks of the entity in the project
In BiCIKL, Plazi will lead the WP6 (JRA-01), is involved in Networking activities, provide in TA the TreatmentBank data preparation service and in VA with its Biodiversity Literature Repository virtual access to the liberated and enhanced data.
Key persons assigned to the project
Dr. Donat Agosti (Male), is founding president of Plazi Verein and director of Plazi GmbH. He promoted in life sciences at ETHZ, Zürich Switzerland, followed by research at the Natural History Museums in London and New York, and at the Jet Propulsion Laboratory, CalTech. Since 1995 he has been involved in building open access, Web based information systems including TreatmentBank and Biodiversity Literature Repository. He has extensive experience in networking at the level of scientists to science managers (Bouchout Declaration on Open Science Knowledge Management). Google Scholar: 6606 citations; H-Index 36.
Dr. Guido Sautter (Male) holds a Master (equivalent) degree in computer science from Karlsruhe Institute of Technology (KIT) as well as Ph.D. in text and data mining, also from Karlsruhe Institute of Technology (KIT). As Plazi's chief software designer and lead developer for TreatmentBank and BLR, he has 15 years experience in developing text and data mining software and related infrastructure (Java, SQL, XML, XSLT, JavaScript), with a focus on biodiversity publications, and also 2 years experience in industrial software development (Java). G. Sautter has published ca. 15 papers on text mining and biodiversity informatics, and co-published several more on cybertaxonomy.
Terry Catapano (Male) is Vice President and Co-Founder of Plazi. He has over 20 years experience designing and implementing strategies, infrastructures, and operations for textual markup, data curation and management, digital preservation, digital repositories. He has worked at New York Public Library, Columbia University, and the University of California, Berkeley. He served on the Editorial Board of the Metadata Encoding and Transmission Standard (METS), as the Chair of the Society of American Archivists Schema Development Team responsible for the Encoded Archival Description (EAD) XML schema, and is lead developer of the TaxPub extension to the Journal Article Tag Suite (JATS). Terry holds a BA in comparative Literature and a MA in English from Columbia University and a MLS from Rutgers University.
Relevant publications
- Chester C, Agosti D, Sautter G, Catapano T, Martens K, Gérard I, Bénichou L (2019) EJT editorial standard for the semantic enhancement of specimen data in taxonomy literature. European Journal of Taxonomy 586: 1-22. doi: 10.5852/ejt.2019.586
- Agosti D, Catapano T, Sautter G, Kishor P, Nielsen L, Ioannidis-Pantopikos A, Bigarella C, Georgiev T, Penev L, Egloff W (2019) Biodiversity Literature Repository (BLR), a repository for FAIR data and publications. Biodiversity Information Science and Standards 3: e37197. doi: 10.3897/biss.3.37197 and 10.5281/zenodo.3534270
- Senderov V, Simov K, Franz N, Stoev P, Catapano T, Agosti D, Sautter G, Morris RA, Penev L (2018) OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system. Journal of Biomedical Semantics 9: 5. doi: 10.1186/s13326-017-0174-5
- Catapano T (2010) TaxPub: An Extension of the NLM/NCBI Journal Publishing DTD for Taxonomic Descriptions. Proceedings of the Journal Article Tag Suite Conference 2010. doi: 10.5281/zenodo.3484285
- Agosti D, Egloff W (2009) Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, [2:53]. doi: 10.1186/1756-0500-2-53
Relevant previous projects
- pro-iBiosphere (FP7 grant 312848): Coordination and policy development in preparation for a European Open Biodiversity Knowledge Management System, addressing Acquisition, Curation, Synthesis, Interoperability and Dissemination, http://www.pro-ibiosphere.eu, timing: 2012-2014.
- EU BON (FP7 grant 308454): EU BON: Building the European Biodiversity Observation Network, http://eubon.eu, timing: 2012-2017.
- ICEDIG (Horizon 2020 grant 777483): Innovation and Consolidation for Large Scale Digitisation of Natural Heritage, https://icedig.eu/. timing: 2018-2020.
- OpenAIRE advance grant. Canonical Person Entity (CPE) stand-off metadata services. timing Feb.-Nov. 2020
Partner 4: Meise Botanic Garden
Description of the legal entity
Meise was founded in the early 18th century and is one of the world's largest botanic gardens. It covers 92ha and contains approximately 4 million preserved and 18,000 living specimens. The collection has a global scope with a focus on Central Africa, Belgium and South-West Europe. There are also important historical collections from Latin America, India and Australia. Notable collectors who have contributed to Meise include Van Heurck, Von Martius, Sieber-von Reichenbach and Crepin. A wide range of taxonomic groups are covered including: vascular plants, lichens, mosses, liverworts, fungi, myxomycetes and algae and active research is carried out on many of these groups. The preserved collection will soon be among an elite group of large herbaria that are completely digitized and available online. The living collections hold particular important examples of Rubiaceae, Balsaminaceae, Euphorbiaceae and Araceae as well as unique collections from the Congo Basin. The seedbank is notable for its collections of wild legumes, and endemic and endangered species from Belgium and Katanga. Meise is also a publisher, both of botanical books and floras, but also the journal Plant Ecology and Evolution. Meise's library is one of the most important botanical collections in Europe. It also collects, catalogues and conserves complementary archives to the herbaria and living collections.
Meise is an active research institution, with programs on the evolution of plants and fungi, taxonomy, ecology, conservation, biodiversity informatics and the spread of invasive species.
Main tasks of the entity in the project
Meise will lead WP4 on improving data infrastructures coordination and interoperability through harmonisation of community policies, standards and guidelines and also act as pillar leader on Joint Research Activities.
Key persons assigned to the project
Dr. Quentin Groom (Male) Ph.D from Essex University and almost 30 year’s experience working in both botanical research and the IT industry. Quentin is a research Scientist and leader of the Biodiversity Informatics team. He has worked on all informatics aspects of the herbarium. His research focuses on the use of information technology in the analysis and dissemination of botanical information. He digitized the Flora of Central Africa, has been involved in many EU and nationally funded projects, including those using and evaluating text mining for data mobilization. He is currently Secretary of the Biodiversity Information Standards (TDWG) organization. In the ICEDIG and Synthesys+ projects he is involved with improving standards for biological specimen data and in improving processes for the digitization of collections. He also has long-term interests in invasive species research and citizen science. He is vice-chair of the Alien-CSI COST Action supporting recording of invasive species through citizen science. Also, the TrIAS project is an open science project on invasive species creating open repeatable workflows from citizen science data to species distribution models and risk assessments.
Mrs Sofie De Smedt (Female) BSc (Hons) (Botany) at Ghent University 1998-2003. Sofie started working at Meise in March 2004 as project leader of the African Plants Initiative project (API). The latter, an international digitisation project funded by the Mellon Foundation, expanded to the Global Plants Initiative (GPI) and resulted in the JSTOR Global Plants website where over 2 million specimen images have been made available. In 2014, she was responsible for the European funded Open-Up project where digital images were made available through the Europeana platform, and for the Linden project, a Flemish funded project where ca. 7000 images were digitised and displayed online through the Meise’s virtual herbarium. Since January 2015, she is the project leader of the DOE! project for the mass digitisation of 1.2 million herbarium specimens at the Botanic Garden Meise over a period of 3 years. This project includes: the updating of the existing digitisation infrastructure; the preparation of the collection for digitisation by an external company (both imaging as databasing); the organisation of internal databasing and quality control; the upgrading of the IT infrastructure; the web portal; and, crowdsourcing
Dr. ir. Mathias Dillen (Male) PhD in Bioscience Engineering, spec. Forest & Nature Management, from the universities of Ghent and Groningen (2013-2017). Mathias has been working at Meise Botanic Garden since early 2018 in the Biodiversity Informatics team, on the ICEDIG project, i.e. the design study for the DiSSCo infrastructure. In the project, he worked on tasks related to collection management systems, image analysis, data cleaning, interoperability, semantic enrichment and on linked open data. He also worked on trials of the Google Cloud Vision API for automated data capture from images, and of the Zenodo infrastructure for mass data and image publishing. Aside from ICEDIG, Mathias has been involved in other informatics projects such as SYNTHESYS+ and LinBi. In addition to research projects, he also contributes to the Garden’s data management, data publication and IT infrastructure, plus development of the Garden’s data portals.
Maarten Trekels (Male) MSc in Physics at the University of Leuven 2004-2009. After his studies, Maarten conducted research at the University of Leuven in nuclear solid state physics. This research was conducted in several large-scale facilities around the globe, using state of the art technology. Inspired by the IT technology, Maarten made the transition to industry. He worked as a system engineer in the aerospace and medical industry, focusing on the design and testing of both hardware and software. In 2019, Maarten joined the biodiversity informatics team at Meise Botanic Garden. He is involved in the SYNTHESYS+ and LinBi projects. His main focus currently on using Wikibase as a tool of biodiversity informatics. Furthermore, Maarten has created a IIIF compliant image server for 2 million herbarium specimen images.
Relevant publications
- Groom Q, Dillen, M, Hardy H, Phillips S, Willemse L, Wu Z (2019) Improved standardization of transcribed digital specimen data. Database, 2019, baz129, https://doi.org/10.1093/database/baz129
- Groom Q, Desmet P, Reyserhove L, Adriaens T, Oldoni D, Vanderhoeven S, … & Simpson A. (2019) Improving Darwin Core for research and management of alien species. Biodiversity Information Science and Standards 3: e38084. https://doi.org/10.3897/biss.3.38084
- Groom, QJ, Adriaens, T., Desmet, P., Simpson, A., De Wever, A., Bazos, I.,... & Helmisaari, H. (2017). Seven recommendations to make your invasive alien species data more useful. Frontiers in Applied Mathematics and Statistics, 3: 13. https://doi.org/10.3389/fams.2017.00013
- Güntsch A, Hyam R, Hagedorn G, Chagnoux S, Röpert D, Casino A,... & Hoffmann J. (2017). Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects (2017) Database, 2017: bax003, https://doi.org/10.1093/database/bax003
- Groom QJ. (2015). Piecing together the biogeographic history of Chenopodium vulvaria L. using botanical literature and collections. PeerJ, 3: e723. https://doi.org/10.7717/peerj.723
Relevant previous project
- pro-iBiosphere (FP7 grant 312848): Coordination and policy development in preparation for a European Open Biodiversity Knowledge Management System, addressing Acquisition, Curation, Synthesis, Interoperability and Dissemination, http://www.pro-ibiosphere.eu, timing: 2012-2014.
- EU BON (FP7 grant 308454): EU BON: Building the European Biodiversity Observation Network, http://eubon.eu, timing: 2012-2017.
- ICEDIG: (H2020 grant 777483) Innovation and consolidation for large scale digitisation of natural heritage (https://www.icedig.eu/). Timing: 2018-2020.
- Synthesys+: Creating an integrated European infrastructure for natural history collections. (https://www.synthesys.info/). Timing: 2019-2022.
- TrIAS (Belgian Science Policy Office) Tracking Invasive Alien Species (https://osf.io/7dpgr/). Timing: 2017-2020.
Equipment involved
MBG maintains a state of the art data portal serving 1.7 million specimen details. This is connected to an image server with more than 2 million images of specimens. A crowdsourcing platform for the transcription of herbarium specimens. A herbarium of 4 million specimens, a living collection of 16,000 taxa, a seed bank, a lab for the preparation of DNA for sequencing and light and electron microscopes.
Partner 5: European Molecular Biology Laboratory (EMBL for ELIXIR Hub and EMBL-EBI)
Description of the legal entity
The European Molecular Biology Laboratory (EMBL) was established as an international research organisation in 1974 and is supported by over twenty countries.
EMBL is a centre of excellence for basic research in molecular biology. It is a distributed international research organisation (27 member states) with its main laboratory in Heidelberg. Research at EMBL emphasizes experimental analysis at multiple levels of biological organisation, from the molecule to the organism. One of EMBL’s five core missions is to develop and make available new technologies for the Life Sciences. Accordingly, EMBL has an extensive success record in developing new biology-driven technologies and providing service-oriented user infrastructures including the Advanced Light Microscopy Facility (ALMF) offering external user access, a high-throughput light microscopy screening facility, the Centre for Bioimage Analysis, the Electron Microscopy Core Facility (EMCF) and the EMBL-EBI data resources. Another core mission of EMBL is to provide advanced training to researchers, which is organised by the EMBL International Centre for Advanced Training (EICAT) and encompasses the EMBL International PhD Programme (EIPP), the EMBL Postdoctoral Programme and the Visitors Programme as well as a large number of workshops, courses and conferences. EMBL maintains strong interactive relationships to the major life science research institutions in Europe and has more than 40 years of experience in integrating European research.
The EMBL European Bioinformatics Institute (EMBL-EBI), the focus of EMBL involvement in the proposed project, is situated in Hinxton, United Kingdom. EMBL-EBI helps scientists realise the potential
of ‘big data’ in biology by exploiting complex information to make discoveries that benefit mankind. It makes public biological data from all over the world freely available to the scientific community via a range
of services and tools, and provides professional training in bioinformatics. EMBL-EBI’s data resources are heavily used by researchers worldwide with EMBL-EBI receiving more than 38 million web page requests daily. The institute has been leading computational biology research since its inception in 1994, with work spanning genomic analysis to systems biology. This includes sophisticated multi-dimensional statistical models for genotype to phenotype associations, single-cell genomics, cancer genomics, phylogenetics, biodiversity, marine biology, infectious disease genomics, deep learning and structural biology. Located on the Wellcome Genome Campus just south of Cambridge in the UK, EMBL-EBI is at the centre of one of the highest concentrations of technical and scientific expertise in the world, with over 650 members of staff representing 66 nationalities.
EMBL-EBI manages a unique set of key biomolecular databases, including the European Nucleotide Archive (ENA; sequence data management, archiving and publication), MGnify (metagenomics analysis resources), UniProt (the Universal Protein Resource), Ensembl and Ensembl Genomes (vertebrate and non- vertebrate genomic resources), InterPro (protein families, domains and motifs) and MetaboLights (metabolic profiling data and identifications).
EMBL-EBI's missions are to provide freely available data and bioinformatics services to all facets of the scientific community in ways that promote scientific progress; to contribute to the advancement of biology through basic investigator-driven research in bioinformatics; to provide advanced bioinformatics training to scientists at all levels, from PhD students to independent investigators; and to help disseminate cutting- edge technologies to industry.
ELIXIR is an inter-governmental treaty based consortium, which builds on existing life sciences data resources and services within Europe to orchestrate the collection, quality control and archiving of large amounts of biological data. It follows a hub-and-nodes model, with a single Hub located at EMBL-EBI in Hinxton, Cambridge, UK (with EMBL providing the legal entity) and a growing number of Nodes located at centres of excellence throughout Europe. ELIXIR is an ESFRI Research Infrastructure in permanent operations phase since 2013. The ELIXIR Consortium Agreement established ELIXIR as an independently governed infrastructure within the legal framework of EMBL. To date, 22 countries plus EMBL have signed the ELIXIR Consortium Agreement: Belgium, Czech Republic, Denmark, Estonia, Finland, France, Germany, Hungary, Ireland, Italy, Israel, Luxembourg, the Netherlands, Norway, Portugal, Slovenia, Spain, Sweden, Switzerland, UK and Greece with Cyprus as an observer. Governments and ministries of ELIXIR Member States are responsible for contributing funding for the ELIXIR Hub and coordinating the scientific community in their country into a national Node that provides services to the ELIXIR community, including data resources, tools, Compute provision, Standards development, Training and support to Industry.
The BiCIKL proposal lead by PENSOFT has been granted the ELIXIR endorsement level on 07/02/2020.
The BiCIKL project will be contributing to the long term sustainability of ELIXIR involving the ELIXIR Hub as one of the beneficiaries and has: 4/4
Committed to supporting Open Science, Open Data and Open Software Principles, ELIXIR’s ELSI guidelines and the ELIXIR Equal Opportunities Strategy.
Confirmed they rely on the ELIXIR Services to implement their scientific activities.
Confirmed they are engaged with the following ELIXIR Nodes to implement their scientific activities (who will play a significant role): EMBL(EMBL-EBI), ELIXIR-CH(SIB).
Committed to providing regular (yearly) feedback on the ELIXIR Services they are using to contribute to the continuous improvement of the ELIXIR Services.
ELIXIR Services are reviewed, approved, provided and sustained by the 22 ELIXIR National Nodes and EMBL.
ELIXIR encourages all users to get in contact with their ELIXIR local Node and the ELIXIR experts when developing and implementing their projects to maximise the benefit of using ELIXIR Services.
Main tasks of the entity in the project
ELIXIR Hub will be the main contact in this grant for administrative, financial and legal matters while ELIXIR Hub & EMBL-EBI, will engage as required in the activities in which they are involved.
ELIXIR Hub will contribute to the executive guidance of the project through leadership of WP 4 and 5 (Transnational and virtual access respectively) and involvement in the Executive Leadership team. The ELIXIR Hub will coordinate the Project call process, as described by WP4, providing transnational access to the new community for named users. This activity will include the running of the call process, administration of reviews and project selection and the distribution of funds to project partners involved in addressing those projects. In addition, the ELIXIR Hub will undertake coordination of virtual access to any users to the partners involved in providing virtual access (WP5). Related activities will also include monitoring of project progression and finalisation of documentation as projects are concluded.
EMBL-EBI will contribute to both responding to providing transnational access to ENA and other EMBL- EBI resources, as required via the project calls in WP4, for instance by assisting with access to data or through provision of data support activities. At a technical level, in WP8, EMBL-EBI will deliver enhancements to the biodata infrastructures, for instance to improve the interoperability between molecular data repositories, museum specimens and the literature.
Key persons assigned to the project
Niklas Blomberg (Male) ELIXIR Director, Project Coordinator. Niklas joined ELIXIR as Director in 2013 following 14 years in the pharmaceutical industry with AstraZeneca. He led the global cheminformatic function from 2006-2011 with responsibility for global delivery of novel computational approaches and external partnerships in screening, and in 2011-2013 led the build of new computational biology/ computational chemistry unit for the AZ inflammatory research area. He has also previously been the Chairman of the board for Bioinformatics for Life Science in Sweden (BILS), Chair of the advisory board for the Swedish e-Science for Cancer prevention and cure project, Advisory board member for the Swedish eScience center and the IMI eTRIKS project. He was co-chair for IMI OpenPHACTS, a project with 24 industrial and academic partners to develop standards and infrastructure for effective data-interoperability across chemistry and biology for drug-discovery research. He is currently coordinating three large infrastructure grants ELIXIR-CONVERGE, CORBEL, EOSC-LIFE.
Jerry Lanfear (Male) ELIXIR Chief Technical Officer. Jerry joined ELIXIR in 2017 after a 20 year career with the Pharmaceutical company, Pfizer. During that time he held a variety of roles including Head of Bioinformatics for the Pfizer Sandwich site (1998-2007), Head of Data Management (2007-2009), Head the Materials and Data Management Centre of Emphasis 2009-2011 and finally Head of IT for the Pfizer Neusentis Unit, 2011-2016. During 2014-2016 he was co-lead of the IMI Data and Knowledge Management Strategic Governance Group. He has extensive experience of direct and matrix leadership of teams and in setting strategy across a variety of disciplines.
Guy Cochrane (Male) leads the European Nucleotide Archive (ENA) at EMBL-EBI. ENA is a platform for the management, sharing, integration and dissemination of sequence data. ENA includes, on the technical side, core databasing infrastructure for the rapid archiving of petabytes of sequence data, the Webin data submission/validation application used by several 1000s of data providers, and sophisticated data discovery and retrieval tools used by many times this number. On the content side, ENA offers extensive public domain data from over 1.5 million species. Providing the European node of the celebrated long-standing International Nucleotide Sequence Database Collaboration, Guy is an authority on large-scale international sequence data sharing across application areas and taxonomies. Guy’s team leads on data coordination in TARA Oceans (marine biodiversity), MGnify (environmental genomics), EMBRIC (blue biotechnology), COMPARE (pathogen surveillance) and FAANG (livestock genomics), inter alia. With a background in cancer research and 16 years of experience in bioinformatics services, Guy has driven numerous developments within sequencing informatics and data coordination, leading the development of data standards (particularly in marine and other environmental omics); global next generation sequence data infrastructure and comprehensive submission, archiving and presentation services; CRAM sequence data compression software; and most recently the data hub and portal system, a portfolio of data coordination tools and services. Current work includes a host of omics data management, coordination and analysis projects and ongoing technology development to scale and extend ENA. In addition to the management of his team of biocurators, bioinformaticians and software engineers, Guy has been involved in board, advisory and steering activities, not least as board member of the Genomics Standards Consortium, Science Committee member of the Global Biodiversity Informatics Facility, Advisory Board member to the German data-intensive environmental science project, GFBio, and Board member of the Species2000/Catalogue of Life biodiversity informatics and taxonomy initiative.
Hannah Hurst (Female) Hannah is an ELIXIR Project Manager. She joined the ELIXIR Hub as Project Manager in January 2018 having spent the previous 6 years at Pfizer as the IMI Project Coordinator where she supported Pfizer with their participation in 40 IMI1 and IMI2 projects and sat on the IMI Operations Working Group. At ELIXIR she also manages the H2020 ELIXIR-CONVERGE project and IMI2 FAIRplus project.
Jeena Rajan (Female) is a Project Lead within the Data Coordination and Archiving team. Jeena leads on data coordination and support for various projects in the biodiversity domain, including UniEuk and the Darwin Tree of Life Project, and was previously involved in the EMBRIC marine project, Jeena has experience in the development, implementation and maintenance of checklists and validation rules within these, around functional annotation and standards-compliant metadata. Jeena also has experience of ensuring data are interoperable between different resources, as well as data compliance to FAIR principles. Jeena is responsible for leading expert ENA helpdesk support for general submissions, validation and curation services to the broad ENA user base.
Relevant publications
- Almeida A, Mitchell AL, Boland M (2019) A new genomic blueprint of the human gut microbiota. Nature 1476-4687. https://doi.org/10.1038/s41586-019-0965-1
- Harrison PW, Alako B, Amid C, Cerdeño-Tárraga A, Cleland I, Holt S, Hussein A, Jayathilaka S, Kay S, Keane T, Leinonen R, Liu X, Martínez-Villacorta J, Milano A, Pakseresht N, Rajan J, Reddy K, Richards E, Rosello M, Silvester N, Smirnov D, Toribio AL, Vijayaraja S, Cochrane G (2018) The European Nucleotide Archive in 2018. Nucleic Acids Res. 2018 Nov. https://doi.org/10.1093/nar/gky1078
- Cook CE, Lopez R, Stroe O, Cochrane G, Brooksbank C, Birney E, Apweiler R (2018) The European Bioinformatics Institute in 2018: tools, infrastructure and training. Nucleic Acids Res. 2018 Nov. https://doi.org/10.1093/nar/gky1124
- Durinx C, McEntyre J, Appel R et al. ( 2017) Identifying ELIXIR Core Data Resources [version 2; referees: 2 approved]. F1000Research, 5(ELIXIR):2422. https://doi.org/10.12688/f1000research.9656.2 Bousfield D, McEntyre J, Velankar S et al. (2016) Patterns of database citation in articles and patents
indicate long-term scientific and industry value of biological data resources [version 1; referees: 3 approved]. F1000Research, 5(ELIXIR):160. https://doi.org/10.12688/f1000research.7911.1
Relevant previous projects
- Project coordinator of ELIXIR-EXCELERATE (2015-2019): an H2020 project (H2020-INFRADEV-1- 2015-1) Fast-track ELIXIR implementation and drive early user exploitation across the life sciences, Grant number: 676559, 2015-2019
- MGP-III (BBSRC: BB/R015228/1) EBI Metagenomics - enabling the reconstruction of microbial populations; to 8/2021.
- EMBRIC (EC: Horizon 2020: 654008) European Marine Biological Research Infrastructure Cluster to promote the Blue Bioeconomy; to 5/2019
- EOSC-Life (824087) EOSC-Life brings together the 13 Biological and Medical ESFRI research infrastructures (BMS RIs) to create an open collaborative space for digital biology. By publishing data and tools in a Europe-wide cloud EOSC-Life aims to bring the capabilities of big science projects to the wider research community. Federated user access (AAI) will allow transnational resource access and authorisation. EOSC-Life establishes a novel access model for the BMS RI: through EOSC scientists would gain direct access to FAIR data and tools in a cloud environment available throughout the European Research Area.
- FAIRplus (802750) This project will develop the guidelines and tools needed to make data FAIR. Through worked examples using IMI and EFPIA data and application and extension of existing methods we willimprove the level of discovery, accessibility, interoperability and reusability of selected IMI and EFPIA data (H2020-JTI-IMI2-2017-12-two-stage)
Equipment involved
ELIXIR unites Europe’s leading life science organisations in managing and safeguarding the increasing volume of data being generated by publicly funded research. It coordinates, integrates and sustains bioinformatics resources across its member states and enables users in academia and industry to access services that are vital for their research. The ELIXIR distributed and virtual Infrastructure brings together the most relevant national bioinformatics resources of 22 countries plus EMBL-EBI that are made accessible as ELIXIR services for Life Science Scientist in Academia and Industry. Currently ELIXIR provides access (via the ELIXIR Nodes) to more than 140 services that are periodically reviewed by external experts and that ELIXIR Nodes have committed to sustain long term. Furthermore, ELIXIR Services are contextualised organised around through alignment with the 5 ELIXIR Platforms (Data, Compute, Tools, Interoperability and Training). Finally, ELIXIR has robust mechanisms to allocate certain Services to Key Service Collections In addition key collection of services have been identified including: (ELIXIR Core Data Resources, ELIXIR Deposition Databases and ELIXIR Recommended Interoperability Resources) being that are recognised by public funders as recommended services for Life Sciences.
EMBL-EBI manages large-scale biological databases, which are available to users via web services available 24/7/365. The demand for rapid access to all publicly available biological databases is constantly growing, as is the volume of biological information held within the databases. To support these needs, EMBL-EBI manages an extensive, high-performance compute infrastructure along with large data-storage farms. EMBL-EBI's state-of-the-art technical architecture is secure, robust, and is distributed in three discrete data centres in different geographical locations to assure long-term security. This gives our data very high protection through redundancy, and provides sufficient capacity and reserve to ensure our management of the rising influx of data and compute requests. As of spring 2018 the main compute farm has 34,000 cores (27,000 high throughput and 7,000 high performance) and the installed disk-based storage capacity is above 200 Petabytes. The internal network has a 100 Gigabit backbone within its data centres and multiple 10 Gigabit connections between data centres and most servers are in these data centres are connected by at least 10 Gigabit networks. EMBL-EBI has two independent 10 Gigabit physical uplinks from the data centres to Janet, Internet2 and Geant (the UK, pan-American and pan-European research networks, respectively).
Partner 6: CERN
Description of the legal entity
CERN, the European Organization for Nuclear Research, is the largest particle physics laboratory in the world. Its flagship accelerator, the LHC, is the world’s most powerful accelerator and is providing research facilities for several thousand high-energy physics researchers from all over the globe. The LHC experiments are designed and constructed by large international collaborations and will collect data over a period of 20 years. These experiments will run up to 1 million computing tasks per day and will generate around 15 petabytes of data per year. This data will be shared with all the participating institutes. The computing capacity required to analyse the data far exceeds the capacity needs of any comparable physics experiments today and relies on the combined resources of some 200 computer centres world- wide. CERN and the particle physics community have chosen grid technology to address the huge data storage and analysis challenge of LHC.
The CERN IT department has about 230 staff, predominantly engineers, who operate one of Europe’s largest research computer centres supporting about 17,000 users. The department has developed leading expertise in large scale data centres and long-standing collaborations with industrial and academic partners in the fields of high performance computing and advanced networking. The CERN IT department has been at the forefront of computing for many years and has coordinated the world’s largest grid project, EGEE (Enabling Grids for E-SciencE), funded under FP6 and FP7. CERN has also prominently contributed to a number of other EU projects aiming at extending the EGEE production grid infrastructure to new geographical areas, to serve new applications domains and to support the grid community. Under FP6 and FP7, the department has been involved in some 20 European Commission-funded projects. CERN is a founding partner of the European Grid Initiative that will provide a sustainable grid infrastructure for Europe’s research communities.
Enshrined in the CERN charter, 60 years ago, is the principle that “... the results of its experimental and theoretical work shall be published or otherwise made generally available” and this has inspired CERN to play a leading role in both European and worldwide Open Access movements, aiming to provide anyone with immediate and free access to the results of scientific research. Combining this Open Science vision with IT innovation, CERN has developed Invenio, an Open Source digital repository platform which powers the CERN Document Server – the CERN institutional repository – and is the basis for INSPIRE – the next-generation High-Energy Physics discipline repository – as well as Zenodo and CERN Open Data Portal. The latter is the platform used by CERN to disseminate and share data coming from the LHC with the world; this data comes along with documentation, software, and virtual machines to enable the easy reproducibility of physics analyses.
Main tasks of the entity in the project
CERN will be participating in WP6 in which improvements in Zenodo workflows to better support the BICIKL project will be implemented. As well, CERN will be part of WP5 to provide Virtual Access to Zenodo infrastructure.
Key persons assigned to the project
Dr. Tim Smith (Male) leads the CERN group that develops, installs, and maintains instances of Invenio, the CERN Open Source Digital Repository system. He is heavily involved in initiatives to drive digital archives at the institutional and subject level and to populate them with content of a broad range of media types. He drove the launch of Zenodo within the OpenAIRE project as an open data service for the long-tail of science. He is jointly responsible for CERN’s Open Source Licence Policy. Prior to these tasks, he led teams responsible for computing farm management and physics data management. He was a work package manager of the EU DataGrid project, the forerunner of EGEE. He holds a PhD in Physics and performed research at the CERN LEP accelerators for 10 years.
José Benito González López (Male) leads the CERN section that is in charge of Invenio, the Digital Repository Framework, and several services that are running on top of it: CERN Document Server – CERN’s institutional repository, Zenodo – open data service for the long-tail of science, CERN Open Data Repositories, Digital Memory projects and also backend development of B2Share (EUDAT service). José is also a very experienced open source software developer and project manager with more than 15 years of experience, many of them devoted to the Open Source Project Indico which is the result of a European Project with the same name.
Alexandros Themistoklis Ioannidis Pantopikos (Male) works at CERN’s IT department and is currently responsible for the Zenodo service, an open data service for the long-tail of science. Alexandros has many years of experience as a software engineer, having led the design and operation of a multitude of production-level commercial and open source software systems. He is also a core maintainer of the open source Invenio framework for digital repositories and has worked on promoting Software as a first-class citizen in the scientific research world.
Relevant publications
- Chen X, Dallmeier-Tiessen S, Dasler R, Feger S, Fokianos P, Gonzalez JB, Hirvonsalo H, Kousidis D, Lavasa A, Mele S, Rodriguez DR, Šimko T, Smith T, Trisovic A, Trzcinska A, Tsanaktsidis I, Zimmermann M, Cranmer K, Heinrich L, Watts G, Hildreth M, Lloret Iglesias L, Lassila-Perini K, Neubert S (2018) Open is not enough. Nature Physics, 15(2):113-119. https://doi.org/10.1038/s41567-018-0342-2
- Nowak K, Nielsen LH, Ioannidis Pantopikos AT (2016) Zenodo, a free and open platform for preserving and sharing research output. Zenodo. May 2016. https://doi.org/10.5281/ZENODO.51902
Relevant previous projectс
- OpenAIRE, OpenAIREplus, OpenAIREConnect, OpenAIREAdvance (H2020, CP-CSA grants): Zenodo was launched within the OpenAIRE project which has provided a wide network of partners promoting the interoperability and discovery of Zenodo’s data. CERN was task leader responsible for developing, operating and maintaining Zenodo.
- HNSciCloud (H2020 grant): Helix Nebula – The Science Cloud. An EC supported initiative on establishing a science cloud through public/private partnerships. CERN has been the coordinator of this European Project.
- EUDAT2020 (H2020 grant): EUDAT2020. CERN was the responsible for Invenio developments to create B2SHARE service
- CRISP (FP7 grant): Cluster of Research Infrastructures for Synergies in Physics. CERN participated in several work packages, including notably the Data Continuum implementation solutions for data lifecycle from the data uptake, through data analysis, up to final publication and dissemination of results.
- EGEE (FP7 grant): Enabling Grids for E-sciencE: the pan-European Grid
Equipment involved
CERN offers temporary prototyping resources configured on virtual machines running as a standard part of CERN’s cloud.
Partner 7: Consortium of European Taxonomic Facilities
Description of the legal entity
CETAF is a European network of Natural History Museums, Botanic Gardens and Research Centers with their associated natural science collections and research expertise. It aims to promote training, research collaborations and understanding in taxonomy and systematic biology as well as to facilitate access to our natural heritage by sharing the information derived from the collections. The CETAF network comprises 63 of the largest taxonomic institutions from 22 European countries. Their collections contain a wide range of specimens including animals, plants, fungi and rocks, and genetic resources which are used for scientific research and exhibitions. Collectively, these collections represent more than 80% of the world’s described species and therefore embody an unprecedented and irreplaceable resource for scientific research across the globe. CETAF member institutions dedicate themselves to both the preservation and promotion of this rich heritage through scientific research, training, public outreach and – in light of this proposal – making the rich data available to scientists all over the world through engagement, harmonisation of policies and procedures, facilitation of skills upgrade and contributing to make scientific literature openly accessible and exploitable. CETAF also is an information exchange platform for researchers from a wide variety of scientific disciplines who carry out pioneering research and develop innovative knowledge exchange pathways. To that end, the CETAF e-Publishing Working Group gathers active members to address the need to support scientific online publishing and open access. Moreover, CETAF collaborates and embraces the development of the European Journal of Taxonomy as a diamond peer-reviewed international scientific journal in descriptive taxonomy, covering the eukaryotic world, with a very high impact factor despite its youth.
From digitisation of collections, to the use of digital media and the stimulation of data sharing, CETAF fosters the development of information services for scientific and public use. For the success of BioKnow, this large network of members as well as associated organisations will prove to be an asset. CETAF will use this network for input into its tasks but also for the dissemination of results and the continuation of activities beyond the projects’ lifetime.
Furthermore, the Muséum Nationale d’Histoire Naturelle (MNHN), Paris will act as a linked third party to CETAF. It is one of the world’s major natural history institutions and contributes to the development and sharing of knowledge on geological and biological diversity, cultures and societies diversity, and the history of planet Earth. The collections of MNHN are recognised as a national research infrastructure for biodiversity studies. They are, quantitatively and qualitatively, in the top three in the world of natural history. They comprise an estimated 67 million specimens and house approximately 790,000 primary types and reference specimens. MNHN, with its unique national status, is also considered as the normal repository for all scientifically significant collections made by other French research bodies. This makes MNHN collections invaluable for conservation management planning and a key research infrastructure to better document global change and all new challenges emerging in the field of biodiversity. The MNHN is currently leading a large-scale digitisation programme, and has made 9.6 million specimens available online. The Museum has been involved in over 50 European Union funded projects under FP7 and H2020, among which SYNTHESYS+, and ICEDIG. It is currently leading large-scale programs on digitization and citizen science. The MNHN also plays a key role in the organisation of the French information system on biodiversity and landscape, being the scientific coordinator of this national project.
The Museum is also a scientific publisher since 1802 and currently publishes 9 international peer-reviewed journals and 9 series of monographs, all related to original scientific results in the fields of the Museum: earth sciences, botany, zoology, biodiversity management, history of sciences, and anthropology.
Additionally - and of special interest in the context of this proposal - the MNHN hosts the European Journal of taxonomy (EJT), a CETAF-endorsed, peer-reviewed journal in descriptive taxonomy. Its content is fully electronic and diamond Open Access, meaning neither authors nor readers have to pay fees. It is published and funded by a consortium of ten European natural history institutions across seven countries. The EJT functions as the entity connecting CETAF and the MNHN for the purpose of this proposal. While fulfilling its role of being primarily a journal publishing taxonomic results, EJT also serves as a model to test further developments for the taxonomic publishing process to meet these challenges. EJT aims at setting up a new production workflow including XMLisation process at the desk-editing level, prior to PDF publication, for producing more confident and pertinent statistics, more accurate and confident data both human and machine readable.
Main tasks of the entity in the project
CETAF will lead WP3 on the engagement and outreach for the Biodiversity Knowledge Hub while also leading the task for a training programme in WP2. CETAF has extensive experience in both domains as it leads or has led similar tasks and work packages in projects such as ICEDIG, BIOTALENT or SYNTHESYS+. The MNHN will mainly be involved in WP6.
Key persons assigned to the project
Ana Casino (Female) (Role: Work Package Leader, WP2) is the Executive Director of CETAF since 2012. In this position, she manages the general secretariat and represents CETAF in projects, actions and initiatives. Among a wide array of responsibilities, she coordinates the vast network of institutions that form CETAF and is one of the 2 deputies coordinators of the Research Infrastructure DiSSCo, included in the ESFRI Roadmap update 2018, as well as a work package and task leader in projects such as ICEDIG, SYNTHESYS+, DiSSCo Prepare and vice chair of the COST Action MOBILISE (Horizon2020). She also actively participates in several of CETAFs’ Working Groups, e.g. on strategy, training, digitization, information science and legislation with a special focus on Access and benefit-sharing, with CETAF being a member of the European ABS Consultation Forum. Before joining CETAF, Ms Casino was the director of the Atlantic Botanic Garden and the Chamber of Commerce in Gijon, Spain.
Karsten Gödderz (Male) works as a Project Coordinator for CETAF in Brussels since March 2016. For CETAF, he works on products and services for the member institutions, supports CETAF initiatives and working groups and is involved in the coordination of several proposals for European research grants as well as their implementation, e.g. as a member of the DiSSCo coordination team or in the ICEDIG, SYNTHESYS+, and DiSSCo Prepare projects. He studied Geography, Political Sciences and Environmental Economics with a focus on sustainability, conservation and biodiversity issues in Bonn, Germany and Warsaw, Poland.
Laura Tilley (Female) works at CETAF since July 2019 and is responsible for coordinating SYNTHESYS+ activities that CETAF leads and is involved in. In addition, she is actively involved in the running of other CETAF initiatives and products, more specifically she assists in coordinating the CETAF Earth Science Group, a role that she has continued from her previous position at the State Natural History Museum Stuttgart, were she was also worked on research projects within the field of Palaeobotany. She has a background in Earth Science, Palaeobotany, Palaeoclimatology and received her PhD from the University of Leeds, UK. She completed her master’s degree in Geology at the University of Leicester, UK.
Marie-Laure Kamatali (Female) is a Project Assistant at CETAF since March 2020. She is working on the implementation of the DiSSCo Prepare project, an H2020- funded project to guide the preparatory phase of the ESFRI research infrastructure DiSSCo, as well developing engagement in all the initiatives CETAF partakes in. She graduated in Communications Studies: New Media and Society in Europe. Her professional journey has created opportunities to work in the non-profit, private and public sectors in various countries within public policy and communications teams with a focus on developing and implementing innovate and empowering narratives.
Céline Cassarino (Female) is the Communications Assistant at CETAF since March 2020. She is involved in all communications activities at CETAF including the promotion of the consortium itself and developing its website as well as its activities in different initiatives and projects. The latter includes e.g. developing communication strategies for the DiSSCo Prepare project. She graduated in Communication Studies. She has collaborated with non-profit organisations in the development of visual design, content and promotion on social media.
Laurence Bénichou (Female) is the Head of the French Museum Science Press at the Muséum National d’Histoire Naturelle in Paris since 2001. She is also the Publication Manager of the European Journal of Taxonomy which she founded in 2011 with a board of European colleagues. Finally, she leads the E-Publishing working group of CETAF (Consortium of European Taxonomic Facilities) since 2015. Expert in the field of scientific publishing for the Ministry of French higher education and research, she specializes in Linnaean Taxonomy publishing, Communication Design and Media. Her research is focused on open access and digital publishing.
Chloé Chester (Female) is the Desk-Editor of the European Journal of Taxonomy, in charge of the implementation of the XML-Based workflow.
Relevant publications
- CETAF, 2018, E-Publishing Guidelines, Recommendations regarding authorship citation and open access.
- CETAF, 2018, Bratislava Declaration on the 2050 Vision of the Convention on Biological Diversity.
- CETAF, 2018, Digital Sequence Information (DSI) of Genetic Resources, Executive Statement.
- Anton Güntsch et al., 2017, Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects. Database (Oxford), 2017 (1).
- CETAF, 2015, Strategy and Strategic Development Plan 2015 – 2025.
Relevant previous projects
CETAF has participated in numerous projects and initiative funded under, FP6, FP7 and Horizon 2020, contributes to the development of global endeavours such as EoL (Encyclopedia of Life), iDigBio, and Species2000, and participates actively as an associated partner or member in relevant organisations (GBIF) and networks (SciCOL, SPNHC, SMBD, GGNB, International Union for Biological Sciences (IUBS)). CETAF has also adopted several other initiatives resulting from previous EU-funded projects to enhance its capacity for collaboration and outreach, such as BHL-Europe. It currently takes part in EU-funded projects such as:
- DiSSCo PREPARE (GA no. 871043). Distributed System of Scientific Collections - Preparatory Phase Project. Project funded under H2020-INFRADEV-2019-2. It will act as the primary vehicle through which DiSSCo RI will raise its overall maturity and set itself in a position to implement its construction programme. The project aims to raise the readiness level of the research infrastructure in five domains (data, science, finance, organisation and technology). Website: https://www.dissco.eu/prepare/; timing: Feb 2020 - Jan 2023.
- SYNTHESYS+. Synthesis of Systematic resources. The H2020 INFRAIA project SYNTHESYS+ brings together the European branches of the global natural science organisations (GBIF, TDWG, GGBN and CETAF) with an unprecedented number of collections, to integrate, innovate and internationalise efforts within the global scientific collections community. Major new developments addressed by SYNTHESYS+ include the delivery of a new virtual access programme, providing digitisation-on-demand services, the construction of a European Loan and Visits System (ELViS), and a new data processing platform (the Specimen Data Refinery), applying cutting-edge artificial intelligence to dramatically speed up the digital mobilisation of natural history collections. Website: https://www.synthesys.info/; timing: Feb 2019 - Jan 2023.
- MOBILISE. This COST action (CA17106) on “Mobilising Data, Experts and Policies in Scientific Collections” aims to build up a cooperative, inclusive, bottom-up and responsive network with active involvement of European stakeholders to support research for biodiversity and geodiversity informatics. MOBILISE will facilitate knowledge and technology transfer across stakeholders, bridging the gaps between biodiversity and geodiversity research and information technology best practices. Website: https://www.mobilise-action.eu/; timing: Oct 2018 - Sep 2022.
- ICEDIG. Innovation and consolidation for large scale digitisation of natural heritage. This H2020 INFRADEV project seeks to refine the design study for DiSSCo, the research infrastructure of natural science collections holding institutions included in the update of the ESFRI Roadmap 2018. Website: https://icedig.eu/; timing: Jan 2018 - Mar 2020.
- BIOTALENT. Talent in biodiversity, Innovative education and new skills to increase engagement in science. Project funded under an ERASMUS+ Call (Action: Strategic Partnerships for Cooperation for innovation and the exchange of good practices). This project developed a blended pilot course that combines e-learning and training in the field to upgrade practitioners in the understanding of biological impact of climate change. Website: http://biotalent.myspecies.info/; timing: Sep 2016 - Aug 2019.
Partner 8: Swiss Institute of Bioinformatics
Description of the legal entity
SIB Swiss Institute of Bioinformatics is an academic not-for-profit foundation, recognized to be of public utility and which federates bioinformatics activities throughout Switzerland. SIB is also a founding member of ELIXIR and it constitutes the only Swiss affiliated institution to ELIXIR.
The Institute includes 60 world class research and service groups that bring together over 750 researchers in the fields of proteomics, transcriptomics, genomics, systems biology, structural bioinformatics, evolutionary bioinformatics, modelling, imaging, literature services, biophysics and population genetics in Basel, Bern, Fribourg, Geneva, Lausanne, Lugano and Zurich. SIB’s expertise is widely appreciated and its infrastructure and bioinformatics resources are used by life science researchers worldwide.
SIB's mission is: Providing world-class core bioinformatics infrastructure and services to the national and international life science community in key fields such as genomics, proteomics and systems biology.
SIB develops and maintains databases of international standing, including UniProtKB/Swiss-Prot (manually curated protein sequence database providing a high level of annotation which receives over half a million unique visitors every month).
SIB develops and supplies software and web platforms for the global life science research community, such as SWISS-MODEL (protein structure homology modelling) and SwissDock (ligand docking), and maintains literature services (MEDLINE mirror, PMC mirror, etc.) to support the curation of biological entities (gene and gene products, pathological functions, small molecules, etc.), SIB manages several bioinformatics Core Facilities which provide bioinformatics and statistical support, as well as services and expertise to life scientists – both in academia and industry, thus enabling them to conduct their research projects and analyze the resulting data. The bioinformatics Core Facilities have numerous international collaborations with International Organizations such as the World Health Organization (WHO) and the Food and Agriculture Organization (FAO).
SIB’s portfolio, which includes some of the world’s major bioinformatics resources, its strong expertise in managing data analysis and services, as well as in evaluating their quality and impact on the life science community, will be essential for several WPs, in particular WP6 & 10. Given the focus of the project on data and literature curation, the SIB Text Mining group, headed by Patrick Ruch, will be SIB’s lead contributor of the project, however other services (Core-IT) of SIB will provide support as well. Patrick Ruch will coordinate WP11’s development to deliver an innovative FAIR Data Place web portal (T10.5) powered with advanced literature search and entity association services and to enhance biodiversity data curation and litterature triage (
Main tasks of the entity in the project
In BiCIKL SIB will lead WP11 and will be also participating in WP1 and WP6.
Key persons assigned to the project
Prof. Dr. Patrick Ruch (Male) is Group Leader and member of the foundation council at the SIB Swiss Institute of Bioinformatics. In parallel, he is also Head of Research at the HEG/HES-SO Geneva (University of Applied Sciences Western Switzerland) and co-leads the ELIXIR Data Platform. Patrick Ruch graduated in both computer science and philosophy (Sorbonne University, Paris). He received his PhD in bioinformatics from the University of Geneva in 2002. He then occupied various corporate and public research positions in Europe (IBM Zürich Research Lab.; EPFL Lausanne) and in the US (National Library of Medicine, National Institutes of Health, Bethesda, MD). In 2008, he became a Professor of Information and Library Sciences at the HEG Geneva. He is the author of more than 100 original scientific publications in peer-reviewed scientific journals. His current research focuses on developing data analysis methods to exploit highly structured semantically- rich data types (Wikipedia/DBPedia, molecular biology databases, ontologies, SPARQL endpoints...) and unstructured content (literature, sequences, patents, …). Bibliometrics: Google Scholar: 3135 citations; H-Index 29.
Dr. Julien Gobeill (Male) is bioinformatician at the Swiss Institute of Bioinformatics (SIB) Text Mining group, and lecturer at the University of Applied Sciences Geneva (HES-SO). He completed his PhD in Computer Sciences at the University of Geneva in 2012, and has authored numerous research papers on text mining (h-index 16 in Google Scholar). Before joining the SIB, he worked in various research institutions including the Bibliomics and Text Mining group of the HES-SO, and the University Hospitals of Geneva. At SIB, he is responsible for the SIB Literature Services (fetching and searching in automatically enriched MEDLINE and PMC entries), and has been involved in several text mining projects, mainly in literature and clinical data.
Dr. Emilie Pasche (Female) is bioinformatician at the Swiss Institute of Bioinformatics (SIB) Text Mining group and research associate at the University of Applied Sciences Geneva (HES-SO). Emilie Pasche is trained in biology (Ba) and bioinformatics (MSc and PhD). She obtained her PhD in 2013 from the University of Geneva. She then worked as postdoc at the University Hospitals of Geneva, before joining the University of Applied Sciences Geneva as a research associate, and later the SIB, as a bioinformatician. Her research focuses on the design of text analytics instruments to support the curation of variants. In particular, she has participated to several TREC Precision Medicine challenges.
Relevant publications
- Gobeill J, Gaudet P, Dopp D, Morrone A, Kahanda I, Hsu YY, Wei CH, Lu Z, Ruch P (2018) Overview of the BioCreative VI text-mining services for Kinome Curation Track. Database (Oxford). 2018 Jan 1. doi: 10.1093/database/bay104.
- Teodoro D, Mottin L, Gobeill J, Gaudinat A, Vachon T, Ruch P (2017) Improving average ranking precision in user searches for biomedical research datasets. Database (Oxford). 2017 Jan 1. doi: 10.1093/database/bax083.
- Mottin L, Pasche E, Gobeill J, Rech de Laval V, Gleizes A, Michel PA, Bairoch A, Gaudet P, Ruch P (2017) Triage by ranking to support the curation of protein interactions. Database (Oxford). 2017 Jan 1. doi: 10.1093/database/bax040.
- Venkatesan A, Kim JH, Talo F, Ide-Smith M, Gobeill J, Carter J, Batista-Navarro R, Ananiadou S, Ruch P, McEntyre J (2017) SciLite: a platform for displaying text-mined annotations as a means to link research articles with biological data. Wellcome Open Res. 2017 Jul 10;1:25 doi: 10.12688/wellcomeopenres.10210.2. eCollection 2016.
- Mottin L, Gobeill J, Pasche E, Michel PA, Cusin I, Gaudet P, Ruch P (2016) neXtA5: accelerating annotation of articles via automated approaches in neXtProt. Database (Oxford). 2016 Jul. pii: baw098. doi: 10.1093/database/baw098.
Relevant previous projects
- Elixir Data Platform - Sustainable Infrastructure to support literature-driven biocuration.
- CINECA - FAIR Research Dataset Management Infrastructure for Life sciences.
- SVIP-O - Swiss Variant Interpretation Platform.
Equipment involved
IT Resources of SIB and location of the resource:
- Two storage clusters with a combined capacity of 2.5 petabytes – Switch Cloud Lausanne
- 70 high-performance servers with a total of 4around 2.000 CPU cores – Switch Cloud Lausanne
- Redundant network connectivity with 2 x 10 GBit/s – Switch Cloud Lausanne
-
Encrypted compute and storage infrastructure – BioMedIT/SIB Genève:
- SAN 90 TB Storage, uncluding 12 TB SSD
- Four compute nodes with 16 cores Intel(R) Xeon(R) CPU E5-4640
Partner 9: University of Tartu
Description of the legal entity
University of Tartu (UTARTU) is Estonia's leading centre of research and training. It preserves the culture of the Estonian people and spearheads the country's reputation in research and provision of higher education. UTARTU belongs to the top 3% of world's best universities. As Estonia's national university, UTARTU stresses the importance of international co-operation and partnerships with reputable research universities all over the world. The robust research potential of the university is evidenced by the fact that the University of Tartu has been invited to join the Coimbra Group, a prestigious club of renowned research universities. As of the end of 2019, the university employed 3,500 people of whom approximately 51% are members of the academic staff (including nearly 200 professors). The proportion of full-time teachers and researchers holding a PhD was approximately 66%. As becomes a respected research university, recent years have witnessed an increase in the number and proportion of UTARTU's academic employees. The number of administrative and support staff providing support services to academic units has remained approximately the same. There are approximately 13,400 students (as of 01.02.2016) studying at the university's four faculties. The number of visiting and international students is over 800. The number of doctoral students is 1,300, with around 100 doctoral defences taking place each year.
University of Tartu Natural History Museum and Botanical Garden (NHM) is a department of UTARTU that works in close cooperation with other departments of the University, offering top-level scientific as well as educational, communicative, technological competence and experiences. NHM is the oldest museum in Estonia, founded in 1803. It has long traditions of sharing knowledge to wide audiences. NHM develops collections in geology, zoology, botany and mycology and offers services to scientists, educators as well as general audiences based on the collections and digital information systems. There are nearly 1,2 million specimens in its collections, the public can access newly renovated exhibition and educational programs, the museum also offers citizen science activities and fosters citizen science networking in Baltic region.
Natural History Museum and Botanical Garden develops e-services for biodiversity data management. The most important service is PlutoF which allows to manage data through a full data lifecycle. PlutoF hosts variety of databases, eg. UNITE Community databases, biobank data, etc.
Main tasks of the entity in the project
In BiCIKL UTARTU will be responsible for the implementation of the data management services which are used for the integration and annotation of biological sample, sequence and literature data.
Key persons assigned to the project
Prof. Urmas Kõljalg (Male) is a professor in mycology of the University of Tartu, Estonia since 2001. He is also serving as a director of the Natural History Museum and Botanical Garden of the same university since 2005. His major research area is biodiversity informatics and molecular taxonomy. During the last eighteen years he has been developing online tools for managing open and linked biodiversity data (http://plutof.ut.ee) and for the metabarcoding of fungi (https://unite.ut.ee). He is leading UNITE Board and Estonian research infrastructure roadmap project NATARC which is developing national e-infrastructure for the natural and earth sciences. Web of Science: 11 745 citations, H-Index 40; Google Scholar: 17354 citations; H-Index 47.
Dr. Kessy Abarenkov (Female) is Senior Researcher at the University of Tartu, Natural History Museum and Botanical Garden. She is specialised in biodiversity informatics, database and system development, and in developing IT tools facilitating the use of fungal barcodes for species identification. She has more than 15 years of experience in developing data management and analysis platforms PlutoF and UNITE. She has published more than 50 research articles and is currently involved as a researcher in the Estonian research infrastructure roadmap project "Natural history archives and information network" (NATARC) and Horizon 2020 project EOSC-Nordic. Web of Science: 6957 citations, H-Index 31; SCOPUS: 7035 citations, H-Index: 32; Google Scholar: 9811 citations; H-Index 35.
Allan Zirk (Male) is Head of Software Development in PlutoF biodiversity platform. His bioinformatics team is currently developing and managing full-fledged PlutoF platform, UNITE services and eElurikkus national portal. He has 15 years of experience working in software development. During this time he has worked in different domains (banking, gaming, biodiversity) in different positions (developer, analyst, project manager, team leader). He has delivered projects, creating tools, and services for scientific community - portals, data gathering, data publishing, mobile applications. His main interest and experience are developing solutions for data-centric workflows using web technologies.
Timo Piirmann (Male) has studied Computer Science in University of Tartu. Timo is a software developer with almost 10 years of experience. Working with PlutoF biodiversity platform his main job has been back-end engineering and API design. He is passionate about keeping everything running smoothly - platform deployment processes and other dev-ops tasks.
Raivo Pöhönen (Male) has studied Computer Science in University of Tartu. Raivo is a software developer with almost 10 years of experience. In his early years he worked on different Java projects. Later, moved to front-end technologies. Besides being a good JavaScript programmer he also knows how to work with people, thus solve tricky user experience (UX) problems.
Filipp Ivanov (Male) has studied Computer Science in University of Tartu. He is a software developer with 10 years of experience. Before joining the biodiversity field he worked for the space industry. Most of his coding is done with front-end technologies. It takes him almost no time being productive in any of the modern front- end frameworks. But his wide skillset allows him to work as a full-stack developer.
Relevant publications
- Nilsson RH, 10 co-authors, Kõljalg U, Abarenkov K (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 47: D259–D264. https://doi.org/10.1093/nar/gky1022
- Kõljalg U, Tedersoo L, Nilsson R.H., Abarenkov K (2016) Digital identifiers for fungal species. Science 352: 1182-1183. https://doi.org/10.1126/science.aaf7115
- Kõljalg U, Nilsson RH, Abarenkov K and 39 co-authors (2013) Towards a unified paradigm for sequence-based identification of Fungi. Molecular Ecology 22: 5271-5277. https://doi.org/10.1111/mec
- Tedersoo L, Abarenkov K, 10 co-authors, Kõljalg U (2011) Tidying up international nucleotide sequence databases: ecological, geographical and sequence quality annotation of ITS sequences of mycorrhizal fungi. PLoS One 6 (9), e24940. https://doi.org/10.1371/journal.pone.0024940
- Abarenkov K and 18 co-authors (2010) PlutoF – a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evolutionary Bioinformatics 6: 189 - 196. https://doi.org/10.4137/EBO.S6271
Relevant previous projects
- EU BON (FP7 grant 308454): EU BON: Building the European Biodiversity Observation Network, http://eubon.eu, timing: 2012-2017.
- NATARC: Estonian research infrastructure roadmap project “Natural history archives and information network”, http://natarc.ut.ee, timing: 2010-2015 and 2016-2021.
- ICEDIG (H2020 grant 777483): Innovation and Consolidation for large-scale Digitisation of natural heritage, https://www.icedig.eu, timing: 2018-2020.
- EOSC-Nordic (H2020 grant 857652): https://www.eosc-nordic.eu, timing: 2019-2022.
- DiSSCo Prepare (H2020 grant 871043): https://www.dissco.eu/prepare/, timing: 2020-2023.
Equipment involved
We have access to Estonian Scientific Computing Infrastructure (ETAIS) HPC cluster with 1032 terabytes of storage and >5000 cores including support for data analysis. From Spring 2020 our data management platform PlutoF will be installed as an EOSC cloud service in ETAIS servers.
Partner 10: LifeWatch ERIC
Description of the legal entity
LifeWatch (LW) ERIC is the e-Science European Research Infrastructure for Biodiversity and Ecosystem Research, a distributed Research e-Infrastructure to advance biodiversity research and to provide major contributions in addressing the big environmental challenges, such as the impact of Climate Change on Earth Biodiversity and Ecosystem Functioning. This goal is achieved by providing access through a single infrastructure to a multitude of sets of data, e-Services and tools enabling the construction and operation of Virtual Research Environments (VREs), which allow the accelerated capture of data with new innovative technologies and knowledge-based decision making-support for the management of biodiversity and ecosystems. LifeWatch ERIC will support virtual access to two of its infrastructures.
Main tasks of the entity in the project
In BiCIKL, the two main tasks that LifeWatch ERIC will lead are the analysis of the technical requirement of users and the Implementation of the Biodiversity Knowledge Hub. Apart from that, LifeWatch ERIC will participate in testing and streamlining interoperability and the alignment of findability, reuse and accessibility. Also in the identification and assistance in putting in place the necessary operational framework, and the identification of the components of the BiKH. LifeWatch ERIC will help in the
translation of the functional diagram of T2.1 and the operational framework of T.2 into educational cloud.
Key persons assigned to the project
Dr. Christos Arvanitidis (Male): (IP) LifeWatch ERIC CEO. PhD in Marine Biology. Former Director of Research, IMBBC, HCMR. Involved in more than 50 research and education projects, coordinated more than 7. He has more than 100 peer- reviewed scientific articles. Guest editor in several Journals; reviewer in more than 45 international peer-reviewed journals. Member of: Board of MARS (European Network of Marine Research Institutes and Stations), Society for the Marine European Biodiversity Data (SMEBD), editorial board of the Word Register of Marine Species (WoRMS), scientific advisory council of the International Polychaete Association (IPA), ICES task group 6 working on the seafloor integrity (EU MSDF), expert pool for the UN World Ocean Assessment and member of the Scientific Council of the Institut Méditerranéen de Biodiversité et d'Ecologie marine et continentale (IMBE), Marseille.
Dr. Eng. Juan Miguel González-Aranda (Male): CTO, LifeWatch ERIC & Director of its ICT Core (Spain Common Facility). ERIC Forum Executive Board representing ENVRI cluster. Telecommunications Engineer and European Doctorate & Master on Industrial Organization & Management, Blockchain Master. He has participated in the EU RI opening-up and developments transfer to other world areas (ENPI, Union for the Mediterranean, EU-LAC es). LW ERIC HoD for the “Global Biodiversity Information Facility”-GBIF- and EU-CELAC RI. Spanish Ministry HoU and Delegate for e-Science & e-Infrastructures European Commission initiatives (DG R&I and DG CONNECT) at e-Infrastructure Reflection Group -eIRG-, European Open Science Cloud (EOSC), Copernicus, EuroHPC Spanish “Sherpa”, Group European Experts Data (GEDE, Biodiversity group) at Research Data Alliance Europe (RDA). Support to the establishment of Knowledge Innovation Communities-European Institute of Innovation & Technology -KIC EIT-, particularly Climate KIC. Deputy Director- Technical of the Doñana Biological Station and Research Technologist-Biodiversity Bioinformatics at the Spanish Council for Scientific Research (CSIC). He has participated in the coordination of FP7 & PF6 projects. Official Rapporteur of the “Euro-Mediterranean Monitoring Committee for the RTD Cooperation” –MoCo–. He has also collaborated with the “Explaining the ST&I Policy Mix: From Policy Rationales to Policy Instruments” EPOM Project-PRIME NoE, ECREINetwork “European clusters and regions for eco-Innovation and eco-Investments network”, and EUMEDIS Network & IST MEDA Plan and OMEN projects. About thirty RDI papers & book chapters’ author.
Dr. Alberto Basset (Male) is the interim Head of the LifeWatch-ERIC Service Centre. He is full professor of Ecology at the University of Salento, with main research interests in biodiversity organisation and ecosystem functioning with a particular focus on aquatic ecosystems. Since 2015, he is the pro tempore President of the European Ecological Federation and of the Euro-Mediterranean Federation of research networks on lagoon ecosystems and is in the board of editors of different international journals. At the national level, he is the Manager of the Joint Research Unit LifeWatch-ITA, which represents the Italian LifeWatch Support Committee. He has coordinated and participated in several EU projects.
Dr. Peter H. van Tienderen (Male) leads the LifeWatch initiative in the Netherlands and is interim member of the LifeWatch Executive Board. He is full professor at the University of Amsterdam and currently Dean of the Faculty of Science. His scientific interest concerns the evolution of biodiversity, and published on a wide range of topics in this field. Applied aspects concern the potential consequences of the introduction of GM crops. His involvement in the development of RI’s is exemplified by, amongst others, two recent LERU papers (Challenges for Biodiversity research in Europe and Four Golden Principles for Enhancing the Quality, Access and Impact of Research Infrastructures).
Dr. Cristina Huertas-Olivares (Female): International Initiatives and Projects, LifeWatch ERIC. PhD in Environmental Sciences, MSc in Water Engineering and a MBA Executive. She has participated in more than 28 R&D projects and 28 Conferences. Co-founder of INORE (International Network of Offshore Renewable Energy). Some positions includes: Head of Marine Energy R&D, Business Development Director, and Responsible for Technology Projects in companies like Abengoa and Ayesa, Head of Environmental Research & Strategy in the Wave Energy Centre, etc. She has been part of scientific committees/ chair of international conferences such as METS, ICOE, WREC. In Spain she has been vice chair of the marine energy Standardisation Committee AENOR and Coordinator of Spanish Maritime Technological Platform.
Relevant publications
- Arvanitidis C, Warwick R, Somerfield PJ, Pavloudi C, Pafilis E, Oulas A, Chatzigeorgiou G, Gerovasileiou V, Patkos T, Bailly N, Hernandez F, Vanhoorne B, Vandepitte L, Appeltans W, Keklikoglou K, Chatzinikolaou E, Michalakis N, Filiopoulou I, Panteri E, Gougousis A, Bravakos P, Christakis C, Kassapidis P, Kotoulas G, Magoulas A (2019) The Collaborative Potential of Research Infrastructures in Addressing Global Scientific Questions. Biodiversity Information Science and Standards 3: e37289W. https://doi.org/10.3897/biss.3.37289
- González-Aranda JM, Koureas D, Addink W, Hirsch T, Arvanitidis C, Sáenz Albanés AJ, Schalk P (2019) Facing e-Biodiversity Challenges Together: GBIO framework-based synergies between DiSSCo and LifeWatch ERIC. Biodiversity Information Science and Standards 3: e38554. https://doi.org/10.3897/biss.3.38554
- Kissling WD, Walls R, Bowser A, Jones MO, Kattge J, Agosti D, Amengual J, Basset A, van Bodegom PM, Cornelissen JHC, Denny AG, Deudero S, Egloff W, Elmendorf SC, Alonso García E, Jones KD, Jones OR, Lavorel S, Lear D, Navarro LM, Pawar S, Pirz R, Rüger N, Sal S, Salguero-Gómez R, Schigel D, Schulz K-S, Skidmore A, Guralnick RP (2018) Towards global data products of Essential Biodiversity Variables on species traits. Nature Ecology & Evolution 2: 1531–1540. https://doi.org/10.1038/s41559-018-0667-3
- González-Aranda Juan Miguel, Rodríguez-Clemente Rafael, Lozano Sebastián (2009) E- Research In International Cooperation Networks In Science & Technology Research”. Book chapter on “EResearch Collaboration: Frameworks, Tools and Techniques. Berlin/Heidelberg: Springer-Verlag. ISBN 978-3-642-12256-9 e-ISBN 978-3-64212257-6, pp. 167-199.
Relevant previous projects
- ERIC FORUM. H2020. INFRASUPP-01-2018-2019 2. ENVRI FAIR. H2020. INFRAEOSC-04-2018-2020.
- EU-CELAC ResInfra. H2020. INFRASUPP-2018-2020
- International Joint and Collaborative Initiative (IJI) Invasive Allien Species. Internal Member States Project (Belgium, Greece, Italy, Portugal, Slovenia, Spain, The Netherlands).
- 2018-2021, BioImaging-GR: A Greek Research Infrastructure for Visualizing and Monitoring Fundamental Biological Processes.2018 – 2022, MOBILISE: Mobilising Data, Policies and Experts in Scientific Collections.
Equipment involved
LifeWatch ERIC is a distributed infrastructure, with common facilities in Spain, Italy and Belgium. It operates servers, HCPs, databases, virtual research environments, etc, in seven EU Member States.
Partner 11: Freie Universitaet Berlin
Description of the legal entity
The Botanic Garden and Botanical Museum Berlin (BGBM) of the Freie Universität Berlin is a centre of biodiversity research in Europe, housing extensive scientific collections of herbarium specimens (about 3.5 million), one of the world’s largest living plants collections, as well as the most complete botanical library in Germany. Research activities are focussed on botanical (incl. algae and fungi) systematics, molecular systematics, genomics and taxonomy. BGBM recognised early the new role of NHMs in the domain of electronic information. Today, the BGBM has a biodiversity informatics research and development group with at present more than 20 staff members (computer scientists, botanists, zoologists, engineers, mathematicians, and technicians). Focal points of R&D activities here are taxonomic information systems, networking of distributed primary biodiversity information and research workflows. The BGBM hosts numerous databases and information systems and is connected with a Gigabit backbone and Gigabit connection to the GEANT network via GWIN.
Main task of the entity in the project
Establish links between specimens and derived sequence data (task lead 7.2). Contribution to thе implementation of a pan-European PID system for Digital Specimens (task 7.1).
Key persons assigned to the project
Anton Güntsch (Male), M.Sc. in Computer Science, Head of Biodiversity Informatics in the Dept. of Research and Biodiversity Informatics at BGBM. Studies of mathematics and information science at the Technical University Berlin. 1994-1997 researcher at the Centre for Logistics and Traffic in Berlin, since 1997 researcher at the Freie Universität Berlin, BGBM. Since 2005 Head of Biodiversity Informatics. Scientific activities: design and implementation of collection and taxonomic databases at meta level and object level; design of cooperative networks of distributed biodiversity information systems; digitisation of living and conserved biological collections. Chair of CETAF Information Science and Technology Commission (ISTC), GBIF-D IT-Commission, DiSSCo Synchronisation Group for Data Standardс and Common Resources.
Gabriele Droege (Female) is the Head of GGBN’s Technical Secretariat hosted at the Botanic Garden and Botanical Museum Berlin, Germany. She co‐led the development of the GGBN Data Standard, GGBN Data Portal as well as the GGBN Document Library and is GGBN's liaison to other biodiversity informatics communities. She has long‐term experience with developing and using global infrastructures for biodiversity informatics with special emphasis on molecular data and linking associated data. Her work at the BGBM also involves the development of internal collection and data workflows as well as the curation of the BGBM DNA bank. She is currently working on her Ph.D. in ornithology at the Zoological Research Museum Alexander Koenig in Bonn, Germany.
Jörg Holetschek (Male) has a diploma in Information Systems Engineering and graduated in 2002. After having worked as a software engineer in Enterprise Resource Planning systems, he became part of the biodiversity informatics team at the Botanic Garden and Botanical Museum Berlin with focus on the design and implementation of biodiversity networks. He was and still is involved in several European projects, including BioCASE (Biological Collections Access Service for Europe), SYNTHESYS I/II/III/+ (Synthesis of Systematic Resources), OpenUp! (Opening up the Natural History Collections for Europeana). In his current position he is responsible for embedding the museum into the landscape of biodiversity networks, for evolving data standards such as ABCD (Access to Biological Collections Data) and for maintaining the BioCASe Provider Software, a data publishing software used in the natural history community. For the Consortium of European Taxonomic Facilities (CETAF) he acts as the GBIF participant node manager.
Relevant Publications
- Berendsohn WG, Güntsch A, Hoffmann N, Kohlbecker A, Luther K & Müller A: (2011) Biodiversity information platforms: From standards to interoperability. In: Smith, V. & Penev, L. (Ed.) e-Infrastructures for data publishing in biodiversity science. ZooKeys 150: 71–87, https://doi.org/10.3897/zookeys.150.2166
- Güntsch A, Berendsohn W (2012) OpenUp! Creating a cross-domain pipeline for natural history data. ZooKeys 209: 47-54. https://doi.org/10.3897/zookeys.209.3179
- Borsch T, Stevens A-D, Häffner E, Güntsch A, Berendsohn WG, Appelhans MS, Barilaro C, Beszteri B, Blattner FR, Bossdorf O, Dalitz H, Dressler S, Duque-Thüs R, Esser H-J, Franzke A, Goetze D, Grein M, Grünert U, Hellwig F, Hentschel J, Hörandl E, Janßen T, Jürgens N, Kadereit G, Karisch T, Koch MA, Müller F, Müller J, Ober D, Porembski S, Poschlod P, Printzen C, Röser M, Sack P, Schlüter P, Schmidt M, Schnittler M, Scholler M, Schultz M, Seeber E, Simmel J, Stiller M, Thiv M, Thüs H, Tkach N, Triebel D, Warnke U, Weibulat T, Wesche K, Yurkov A, Zizka G (2020) A complete digitization of German herbaria is possible, sensible and should be started now. Research Ideas and Outcomes 6: e50675. https://doi.org/10.3897/rio.6.e50675
- Droege G, Barker K, Seberg O, Coddington, Jonathan, Benson, E, Berendsohn WG, Bunk B, Butler C, Cawsey EM, Deck J, Döring M, Flemons P, Gemeinholzer B, Güntsch A, Hollowell, Thomas H, Kelbert P, Kostadinov I, Kottmann R, Lawlor RT, Lyal C, Mackenzie-Dodds J, Meyer C, Mulcahy D, Nussbeck SY, O'Tuama é, et al. (2016) The Global Genome Biodiversity Network (GGBN) Data Standard specification. Database, 2016: 1-11. https://doi.org/10.1093/database/baw125
- Güntsch A, Hyam R, Hagedorn G, Chagnoux S, Röpert D, Casino A, Droege G, Glöckler F, Gödderz K, Groom Q, Hoffmann J, Holleman A, Kempa M, Koivula H, Marhold K, Nicolson N, Smith VS, Triebel D (2017) Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects. Database (Oxford) 2017; 2017 (1): bax003. https://doi.org/10.1093/database/bax003
Relevant Projects
- OpenUp! (FP7 ICT-PSP-CIP-grant 270890) OpenUp! (Opening up the Natural History Heritage for Europeana). 23 Partners from 12 countries. timing: 1. 3. 2011-30. 4. 2014.
- ViBRANT (FP7-ICT-2007-1, FP7 grant 261532) ViBRANT - Supporting biodiversity research communities, timing: 01.12.2010-30.11.2013.
- BioVeL (EU, INFRA-2011-1.2.1, FP7 grant 283359) BioVeL - Virtual e-Laboratory, meets the needs of Europe’s Biodiversity Science research community with tools for pipelining data and analysis into efficient workflows, timing: 01.09.2011-31.08.2014.
- Pro-iBiosphere (FP7 grant 312848): Pro-iBiosphere - Coordination and policy development in preparation for a European Open Biodiversity Knowledge Management System, addressing Acquisition, Curation, Synthesis, Interoperability and Dissemination, timing: 01.09.2012 - 31.08.2014.
- BioCASE (EVR1-CT-2001-40017) BioCASE - A Biological Collection Access Service for Europe. 35 Partners from 30 countries, timing: 1.11.2001-31.01.2005.
Partner 12: Global Biodiversity Information Facility
Description of the legal entity
GBIF - the Global Biodiversity Information Facility - is an international network and research infrastructure funded by the world’s governments and aimed at providing anyone, anywhere, open access to data about all types of life on Earth.
Established under a non-binding Memorandum of Understanding, GBIF is coordinated through a Secretariat based in Copenhagen established as a juridical body under Danish law through a country host agreement. The GBIF network of participating countries and organizations, working through participant nodes, provides data-holding institutions around the world with common standards and open-source tools that enable them to share information about where and when species have been recorded.
As of February 2020, the global GBIF network comprises 59 country participants, 41 other associate participants and affiliates (most of them international organizations), and more than 1,500 data-publishing institutions. GBIF is uniquely qualified to coordinate the internationalization efforts as the coordinator of the alliance for biodiversity knowledge, which arose from two Global Biodiversity Informatics Conferences that produced the Global Biodiversity Information Outlook (GBIO) and other resulting GBIO documents. The alliance is open worldwide to all institutions, agencies, organizations, researchers and communities working to measure and monitor biodiversity or dependent on accurate information on biodiversity. Its aim is to align resources and investments in biodiversity informatics and deliver current, accurate and comprehensive information and knowledge on the world's biodiversity in support of a sustainable future for science and society.
In recent years, GBIF has been actively involved in the planning, management and execution of numerous European projects, including SYNTHESYS+, DiSSCo PREPARE, EU BON and BioFresh. Since 2015, GBIF has also led the Biodiversity Information for Development (BID) programme, funded by EU DEVCO to increase availability and use of open biodiversity data across the countries of sub-Saharan Africa, the Caribbean and the Pacific Islands.
The 28-person GBIF Secretariat is currently organized into four teams:
-
Participation and Engagement is responsible for operating the network of Participants and publishers, recruiting new members and enhancing the capacity of current ones.
-
Data Products is responsible for the quality and scientific value of the integrated data products produced by the GBIF network.
-
Informatics is responsible for data management, software development and the overall operation of the GBIF infrastructure.
Administration is responsible for maintaining both the network and the Secretariat’s underlying operations and processes.
Main task of the entity in the project
GBIF leads WP 2 including tasks 2.1 and 2.2. GBIF is also involved in WP10. The main work focuses on defining & co-designing the Biodiversity Knowledge Hub (BKH) and operational training.
Key persons assigned to the project
Kyle Copas (Male) is Communications Manager for the Global Biodiversity Information Facility. He leads outreach for and to the GBIF network and its stakeholders and has represented GBIF widely across the global citizen science community. He currently coordinates GBIF’s activities for the alliance for biodiversity knowledge, after serving as one of the organizers of the 2nd Global Biodiversity Informatics Conference and co-author of the publication outlining the vision for the alliance. He previously played several roles at NatureServe, a U.S.-based conservation non-profit (and GBIF participant). He earned his BA from Wabash College.
Dr. Joseph Miller (Male) is the Executive Secretary of the Global Biodiversity Information Facility. His research interests include plant systematics, biogeography and biodiversity informatics, focusing mainly on the Australian flora. Miller spent six years at the US National Science Foundation where he promoted international research and managed biodiversity science programs. From 2008-2013, Miller was a senior research scientist at the Australian National Herbarium in Canberra. He has a PhD from the University of Wisconsin. Miller has 93 peer-reviewed publications, Google Scholar: 4188 citations; H-Index 38.
Relevant Publications
-
GBIF.org platform and data index: central index of 1.4 billion species occurrence records, drawn from more than 50,000 datasets contributed by 1,500-plus institutions worldwide, with free and open search, API and download access supported by a central backbone taxonomy based on the Catalogue of Life (CoL partnership): https://www.gbif.org
-
Hobern D, Baptiste B, Copas K, Guralnick R, Hahn A, van Huis E, Kim E, McGeoch M, Naicker I, Navarro L, Noesgaard D, Price M, Rodrigues A, Schigel D, Sheffield C, Wieczorek J (2019) Connecting data and expertise: a new alliance for biodiversity knowledge. Biodiversity Data Journal 7: e33679. https://doi.org/10.3897/BDJ.7.e33679
-
Integrated Publishing Toolkit (IPT), https://www.gbif.org/ipt A free open source software tool used to publish and share biodiversity datasets through the GBIF network; latest release: 2.3.5. Currently 278 installations in 73 countries.
-
The trend of occurrence records mobilised through GBIF over time is recognised by the Biodiversity Indicators Partnership as an indicator of progress towards Aichi Target 19 under the CBD’s Strategic Plan for Biodiversity 2011-2020
-
Chandler M, See L, Copas K, Bonde AMZ, Claramunt B, Danielsen F, Legind JK, Masinde S, Miller-Rushing AJ, Newman G, Rosemartin A & Turak A (2017) Contribution of citizen science towards international biodiversity monitoring. Biological Conservation 213(B): 280-294. https://doi.org/10.1016/j.biocon.2016.09.004
Relevant Projects
- Partnerships and affiliations beyond the GBIF network itself include observer status in the CBD (Convention on Biological Diversity), IPCC (Intergovernmental Panel on Climate Change), IPBES (Intergovernmental Platform on Biodiversity and Ecosystem Services), IUCN (International Union for the Conservation of Nature) and membership in the ICSU World Data System, ECSA (European Citizen Science Association) as well as partnership in DataOne and the Global Partnership for Plant Conservation, among others.
- BID: Biodiversity Information for Development, lead [2015-2019]. A multi-year million programme funded by EU DG DEVCO and led by GBIF, with the aim of increasing the amount of biodiversity information available in the ‘ACP’ nations of sub-Saharan Africa, the Caribbean and the Pacific. Timing: 2015-present.
- SYNTHESYS+ (H2020 grant 871043): Provide guidance and oversight for integrating international participation of global scientific collections community. Timing: 2019-2023.
- DiSSCo PREPARE: Provide guidance and oversight for integrating international participation of global scientific collections community. Timing: 2020-2023.
- EU BON (FP7 grant 308454): Building the European Biodiversity Observation Network, member.. Coordination of the review and guidelines for using standards in order to improve interoperability. Timing: 2012-2017.
Equipment involved
GBIF operates a computing cluster to power our core occurrence store and index, plus database servers, virtual machine hosts and storage systems. Servers are housed in a secure University of Copenhagen datacentre. The servers share a redundant 40Gb/s uplink to the University of Copenhagen network, which links to the Danish national research network (multiple 100GB/s), and thus to GÉANT, the pan-European research and education network (multiple 100GB/s).
Partner 13: Species 2000
Description of the legal entity
Species 2000 is an autonomous federation of taxonomic database custodians, involving taxonomists throughout the world. Our goal is to collate a uniform and validated index to the world's known species (plants, animals, fungi and microbes). Species 2000 is registered as a not-for-profit company limited by guarantee (registered in England No. 3479405) but is currently shifting its registration to the Netherlands.
Species 2000 began as a joint programme between CODATA (International Council for Science: Committee on Data for Science and Technology), IUBS (International Union of Biological Sciences) and the IUMS (International Union of Microbiological Societies) in the early 1990's. In 1996 eighteen taxonomic database organisations agreed to convert Species 2000 into a legal entity as the vehicle for developing the global Species 2000 programme. It is an associate participant in the Global Biodiversity Information Facility (GBIF); a data provider to EC LifeWatch ; and is recognised by the United Nations Environment Program (UNEP) and the Convention on Biological Diversity (CBD).
Species 2000 is a partner of the Consortium of European Taxonomic Facilities (CETAF) and works with their community on the construction of an authoritative taxonomic index of species: Catalogue of Life.
Species 2000 has a distributed Secretariat: the administrative office and staff are hosted and sponsored by Naturalis Biodiversity Center in the Netherlands and the Editorial Office is hosted and sponsored by the Illinois Natural History Survey in the USA, and informatics support is managed by the GBIF Secretariat in Denmark.
Species 2000 partners with the Integrated Taxonomic Information System (ITIS) to deliver the Catalogue of Life (CoL). CoL is the most comprehensive and authoritative global index of species currently available. It consists of a single integrated species checklist and taxonomic hierarchy. The Catalogue holds essential information on the names, relationships and distributions of over 1.9 million species. This figure continues to rise as information is compiled from diverse sources around the world. More information about the collaboration between Species 2000 and ITIS can be found under Organisation on the Catalogue of Life Website.
The Catalogue of Life 2019 Annual Checklist is only available online and no longer on dvd-rom as in earlier years since this medium is becoming obsolete. The Annual Checklist is also available as Darwin Core Archive.
The data infrastructure and production environment of the Catalogue of Life is currently being redeveloped in the context of the COL+ project. Focus during phase one of this development is on streamlining the ingestion and processing of data from providers to be included in the COL, as well as on enhancing the services to institutional users such as GBIF, EOL, BHL, BOLD, LifeWatch, DiSSCo and WFO. A second phase of the COL+ will focus on empowering the taxonomic community to feed data directly into the COL and be in control of that process.
Main tasks of the entity in the project
Species 2000 will deliver high-quality virtual access to the taxonomic framework for use throughout the BiCIKL project and strengthen linkages with the taxonomic community and taxonomic publishers to ensure the quality of this framework and its trustworthiness.
Key persons assigned to the project
Dr Peter Schalk (Male) is Deputy Director, Naturalis Biodiversity Center, The Netherlands and the Executive Secretary of Species 2000. He holds a PhD in marine biology, specialized in taxonomy, ecology, and biodiversity informatics. He worked as scientific researcher at the University of Amsterdam, Netherlands Institute for Sea Research, the Alfred Wegener Institute for Polar Research, and as managing director at ETI Biodiversity Center. Since 2013 he has been Deputy Director at Naturalis Biodiversity Center, responsible for the museum, education, business development and international collaboration. Peter is also the Executive Secretary of Species 2000, the legal body behind the Catalogue of Life, of which he was one of the founders. From 2014-18 he was Governing Board Chair of the Global Biodiversity Information Facility (GBIF), and currently part of the Executive Committee in his role as Budget Committee Chair.
Dr Olaf Bánki (Male) is coordinator Catalogue of Life Plus initiative and ESFRI internationalisation. As biodiversity information specialist, he has worked for several years for the Global Biodiversity Information Facility Secretariat, being responsible for engaging GBIF member countries and organisations in serving biodiversity information for science and societal solutions. Olaf assisted the coordination of the preparatory phase of the ESFRI LifeWatch e-infrastructure for biodiversity and ecosystem research. For Global Earth Observations - Biodiversity Observation Network he was a member of the steering committee for the biodiversity data interoperability. Olaf is experienced in fundamental tropical plant biodiversity research (H-index = 14; RG score = 26.41), environmental impact assessment, nature conservation and species red list assessments, and international biodiversity policy influence. Currently Olaf is a member of the IUCN Commission on Ecosystem
management, the chair of the Van Tienhoven Foundation for international nature protection, and a member of the Catalogue of Life global team.
Donald Hobern (Male) is International Engagement Officer for Species 2000 and Executive Secretary for the International Barcode of Life Consortium. Between 2012 and 2019, he served as GBIF Executive Secretary and Director of the GBIF Secretariat. Previously, he held positions as Director of the Atlas of Living Australia (2008-2012), Chair of the TDWG Executive Committee (2008-2010), GBIF Programme Officer for Data Access and Database Interoperability and GBIF Deputy Director for Informatics (2002-2007). He holds an MA in Classics and worked from 1987 to 2002 as a software developer and then web architect for IBM UK and IBM New Zealand.
Mil de Reus (Female) is a Management and Project Assistant at Naturalis and Species 2000. She has a BA in Business Economics and worked as an Office Manager for ETI (1996-2013). She now is a Management Assistant/Project Assistant at Naturalis/Species 2000 (from 2013).
Relevant publications
-
Banki O, Hobern D, Döring M, Remsen D (2019) Catalogue of Life Plus: A collaborative project to complete the checklist of the world's species. Biodiversity Information Science and Standards 3: e37652. https://doi.org/10.3897/biss.3.37652
-
Hobern D, Banki O, Döring M, Remsen D (2019) Supporting 21st Century Taxonomy and Society Through Collaborative Cataloguing of the World's Species. Biodiversity Information Science and Standards 3: e37325. https://doi.org/10.3897/biss.3.37325
-
Ower G, Roskov Y (2019) The Catalogue of Life: Assembling data into a global taxonomic checklist. Biodiversity Information Science and Standards 3: e37221. https://doi.org/10.3897/biss.3.37221
-
Döring M, Ower GD (2019) The Catalogue of Life Data Package - A new format for exchanging nomenclatural and taxonomic information. Biodiversity Information Science and Standards 3: e38771. https://doi.org/10.3897/biss.3.38771
-
Bisby FA, Roskov YR (2010) The Catalogue of Life: towards an integrative taxonomic backbone for biodiversity, in Pier Luigi Nimis and Régine Vignes Lebbe (eds.): “Tools for Identifying Biodiversity: Progress and Problems. Proceedings of the International Congress, Paris, September 20-22, 2010”, Trieste, EUT Edizioni Università di Trieste, pp. 37-42.
Relevant previous projects
- OpenUp! (2011-2014) - Opening up the Natural History Heritage for Europeana - EU contribution € 23,808
- EUBrazilOpenBio (2011-2013) - EU-Brazil Open Data and Cloud Computing e-Infrastructure for Biodiversity.
- i4Life (2010-2013) - Indexing for Life.
- BHL-Europe (2009-2012) - Biodiversity Heritage Library for Europe.
- 4D4Life (2009-2012) - Distributed Dynamic Diversity Databases for Life.
Partner 14: Stichting International Working Group on Taxonomic Databases (TDWG Europe)
Description of the legal entity
TDWG is a not for profit scientific and educational foundation that is affiliated with the International Union of Biological Sciences. TDWG was formed to establish international collaboration among biodiversity information resources. It promotes the integration and dissemination of biodiversity information by developing data exchange standards and by serving as a forum for discussion and professional development. Networks of data publishers and integrators on every continent, as well as globally, use TDWG’s most popular standards, the Darwin Core and ABCD, to create the most comprehensive repositories of primary biodiversity data. The largest of these is operated by the Global Biodiversity Information Facility (GBIF) is approaching one billion records. TDWG is an open, bottom- up, organisation of individual and institutional members, who sustain the organisation with modest membership dues. Members self-assemble into interest groups to create standards and best practices. While the two most widely used standards are now more than 15 years old, they have evolved to adapt to the ever- changing technologies of the Internet, and newer interest groups are developing new standards to encompass new domains. Since its founding in 1985, TDWG has convened an international conference every year, and since 2003 these conferences have drawn an average of more than 150 participants. The TDWG conference is consistently the best place for biologists, IT experts, and computer scientists to present and assess the state of art in Biodiversity Information Science. In 2017, TDWG engaged Pensoft publishers to produce our proceedings in a new journal, Biodiversity Information Science and Standards (BISS).
Main tasks of the entity in the project
Supporting standards development, but also supporting interest and task groups. TDWG will ensure that standards development will occur in a global context and will connect the project to partners in other continents.
Key persons assigned to the project
Patricia Mergen (Female) is currently liaison officer at Meise Botanic Garden and member of the Biodiversity Information Standards (TDWG) executive where she is the chair of the Time and Place Committee for organizing the TDWG annual meetings and outreach to the participants. She has been managing EU projects for over 20 years and is specialized in data standards, numerical ecology and biomonitoring. She is a promoter in Flanders of the infrastructure DiSSCo on scientific collections, involved in the DiSSCo linked EU projects and sits as an expert in the EOSC sustainability Working Group to channel the implementation of the Open Science Cloud and application of FAIR data in Europe. She is mentor and training provider on Biodiversity Information topics within GBIF and training schools of the COST Action of DiSSCo (Mobilise).
Deborah Paul (Female) is currently Digitization and Workforce Development Manager for the iDigBio Project and Vice-chair for the International Working Group on Taxonomic Databases (TDWG). She will begin a two-year term as TDWG Chair in 2021. Research interests center around international collaboration and capacity development needs to foster biocollections digitization and data use. She recently served as an expert external advisor for the ICEDIG design study. Presently, Deborah is co-leading development of a new data standard for collections metadata to support data sharing and aggregation for biological and geological collections.
Stan Blum (Male) has been the principal analyst or a contributor to the design of scientific databases for the National Museum of Natural History (Smithsonian), Museum of Vertebrate Zoology (UC Berkeley), University of Kansas (“Specify”), and the California Academy of Sciences. He has been an active participant in TDWG since 1998, and served as Chair (2001-2003) and Treasurer (2004-2009). The dominant theme in his career has been the integration of information across organizations and disciplines to support biodiversity science. Currently he serves as Administrator of the TDWG Secretariat.
Relevant publications
-
Baskauf S, Wieczorek J, Blum S, Morris RA, Rees J, Sachs J, & Whitbread G (2017). TDWG Vocabulary Maintenance Specification.
-
James SA, Soltis PS, Belbin L, Chapman AD, Nelson G, Paul DL, & Collins M (2018) Herbarium data: Global biodiversity and societal botanical needs for novel research. Applications in plant sciences, 6(2), e1024. https://doi.org/10.1002/aps3.1024
-
Thessen AE, Woodburn M, Koureas D, Paul D, Conlon M, Shorthouse DP, & Ramdeen S (2019) Proper Attribution for Curation and Maintenance of Research Collections: Metadata Recommendations of the RDA/TDWG Working Group. Data Science Journal, 18(1). https://doi.org/10.5334/dsj-2019-054
-
Nelson G, Paul D, Riccardi G, Mast A (2012) Five task clusters that enable efficient and effective digitization of biological collections. ZooKeys 209: 19-45. https://doi.org/10.3897/zookeys.209.3135
-
Seltmann K, Lafia S, Paul D, James S, Bloom D, Rios N, Ellis S, Farrell U, Utrup J, Yost M, Davis E, Emery R, Motz G, Kimmig J, Shirey V, Sandall E, Park D, Tyrrell C, Thackurdeen R, Collins M, O'Leary V, Prestridge H, Evelyn C, Nyberg B (2018) Georeferencing for Research Use (GRU): An integrated geospatial training paradigm for biocollections researchers and data providers. Research Ideas and Outcomes 4: e32449. https://doi.org/10.3897/rio.4.e32449
Relevant previous projects
- Synthesys+: Creating an integrated European infrastructure for natural history collections. (https://www.synthesys.info/). Timing: 2019-2022.
Third parties involved in the project (including use of third party resources)
Muséum Nationale d’Histoire Naturelle (MNHN), Paris will act as a linked third party to CETAF. It is one of the world’s major natural history institutions and contributes to the development and sharing of knowledge on geological and biological diversity, cultures and societies diversity, and the history of planet Earth. The collections of MNHN are recognised as a national research infrastructure for biodiversity studies. They are, quantitatively and qualitatively, in the top three in the world of natural history. They comprise an estimated 67 million specimens and house approximately 790,000 primary types and reference specimens. MNHN, with its unique national status, is also considered as the normal repository for all scientifically significant collections made by other French research bodies. This makes MNHN collections invaluable for conservation management planning and a key research infrastructure to better document global change and all new challenges emerging in the field of biodiversity. The MNHN is currently leading a large-scale digitisation programme, and has made 9.6 million specimens available online. The Museum has been involved in over 50 European Union funded projects under FP7 and H2020, among which SYNTHESYS+, and ICEDIG. It is currently leading large-scale programs on digitization and citizen science. The MNHN also plays a key role in the organisation of the French information system on biodiversity and landscape, being the scientific coordinator of this national project.
The Museum is also a scientific publisher since 1802 and currently publishes 9 international peer-reviewed journals and 9 series of monographs, all related to original scientific results in the fields of the Museum: earth sciences, botany, zoology, biodiversity management, history of sciences, and anthropology. Additionally - and of special interest in the context of this proposal - the MNHN hosts the European Journal of taxonomy (EJT), a CETAF-endorsed, peer-reviewed journal in descriptive taxonomy. Its content is fully electronic and diamond Open Access, meaning neither authors nor readers have to pay fees. It is published and funded by a consortium of ten European natural history institutions across seven countries. The EJT functions as the entity connecting CETAF and the MNHN for the purpose of this proposal. While fulfilling its role of being primarily a journal publishing taxonomic results, EJT also serves as a model to test further developments for the taxonomic publishing process to meet these challenges. EJT aims at setting up a new production workflow including XMLisation process at the desk-editing level, prior to PDF publication, for producing more confident and pertinent statistics, more accurate and confident data both human and machine readable.
In BiCIKL, the MNHN will mainly be involved in WP6 JRA-01 Liberation of data from literature, next- generation semantic publishing and delivery of FAIR data and will contribute to WP2 NA-2: Building the Biodiversity Knowledge Hub (BKH) and operational training.
Ethics and security
Ethics
All BiCIKL partners are committed to the highest ethical standards. The coordinator, Pensoft, has passed GDPR training and certification. We confirm that we have taken into account all ethics issues and will complete the ethics self-assessment and attach the required documents. BiCIKL addresses some ethical issues in this chapter. Within the project framework the collection, processing and storage of potentially sensitive data will be restricted to the contact data of the individuals representing the beneficiaries, associated partners and a range of stakeholders. No other personal data than contact data (i.e. name, address, e-mail, telephone) will be collected in the framework of the present project. A dedicated platform for handling personal data will be created for applicants for TA. Its collection and associated processing procedures will comply with national and EU legislation (in particular the General Data Protection Regulation (GDPR) (EU) 2016/679. All information containing personal data will be collated in a secure database that will be stored on secure, password/token-protected servers within the EU. Most of the organisations in the consortium will appoint a Data Protection Officer (DPO) to comply with the GDPR with respect to contact details to the research participants. Dedicated consent forms will be created and applied in the procedures for TA and VA. Furthermore, security measures will be implemented at technical level to prevent unauthorised access to personal data, including anonymisation
/pseudonymisation techniques. Data Management Plan (DMP) will be prepared at the start of the project to describe all the types of data, licenses and ownership and guarantee its proper management and data processing operations throughout the project duration.
Specifically, BiCIKL will ensure that:
-
Personal data will be processed legally and fairly.
-
Data will be collected for explicit and legitimate purposes and used accordingly.
-
Data will be adequate, relevant and restricted to the purposes for which it is collected and processed.
-
Data will be accurate and updated where necessary.
-
Data subjects can rectify, remove or block incorrect data.
-
Data that identifies individuals (personal data) will not be kept any longer than necessary.
-
Personal data will be protected against accidental or unlawful destruction, loss, alteration and disclo- sure, particularly when processing involves data transmission over networks. Appropriate security measures will be implemented.
The BiCIKL consortium confirms that the ethical standards and guidelines of Horizon 2020 will be rigorously applied, regardless of the country in which the research is carried out. All BiCIKL beneficiaries are legally based in the EU and will, without exception, adhere to Horizon 2020 ethical rules and data security regulations. Again, BiCIKL contact databases will be stored exclusively within the EU at all times. BiCIKL will request informed consent from the representatives taking part in the meetings regarding the storage of their contact data, images, and of their statements within the minutes of these meetings even if these statements are likely to refer to discussions of confidential and potentially controversial issues. For this purpose, a common BiCIKL Informed Consent Document will be produced and agreed upon among the individuals affected. The BiCIKL management structure, that includes the election of an Ombudsperson at the first GA meeting, is constructed to effectively monitor and ensure that good ethics are upheld in the project. The Equality and diversity champion will oversee the adherence to all aspects of equal treatment, including the gender dimension.
Security
BiCIKL will not involve activities or produce results raising security issues. The project will not use EU-classified information' as background or results".
Funding program
H2020-INFRAIA-2020-1: Integrating Activities for Starting Communities
Grant title
Biodiversity Community Integrated Knowledge Library (BiCIKL)
Hosting institution
Pensoft Publishers
Conflicts of interest
References
- Biodiversity data are out of local taxonomists' reach.Nature439(7075):392‑392. https://doi.org/10.1038/439392a
- Taxonomic information exchange and copyright: the Plazi approach.BMC Research Notes2(1). https://doi.org/10.1186/1756-0500-2-53
- The Plazi Workflow: The PDF prison break for biodiversity data.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37046
- A plea for digital reference collections and other science‐based digitization initiatives in taxonomy:Sepsidnet as exemplar.Systematic Entomology38(3):637‑644. https://doi.org/10.1111/syen.12015
- Approaches to estimating the universe of natural history collections data.Biodiversity Informatics7(2). https://doi.org/10.17161/bi.v7i2.3991
- Biodiversity into your hands - A call for a virtual global natural history ‘metacollection’.Frontiers in Zoology10(1). https://doi.org/10.1186/1742-9994-10-55
- The Biodiversity Informatics Landscape: Elements, Connections and Opportunities.Research Ideas and Outcomes3https://doi.org/10.3897/rio.3.e14059
- Utilizing online resources for taxonomy: a cybercatalog of Afrotropical apiocerid flies (Insecta: Diptera: Apioceridae).Biodiversity Data Journal3https://doi.org/10.3897/bdj.3.e5707
-
She Figures 2015 - Gender in Research and Innovation. Directorate-General for Research and Innovation.URL: https://tinyurl.com/she-figures-2015
- The Earth BioGenome project: opportunities and challenges for plant genomics and conservation.The Plant Journal102(2):222‑229. https://doi.org/10.1111/tpj.14631
- Overview of the BioCreative VI text-mining services for Kinome Curation Track.Database2018https://doi.org/10.1093/database/bay104
- Community Next Steps for Making Globally Unique Identifiers Work for Biocollections Data.ZooKeys494:133‑154. https://doi.org/10.3897/zookeys.494.9352
- A decadal view of biodiversity informatics: challenges and priorities.BMC Ecology13(1). https://doi.org/10.1186/1472-6785-13-16
-
Global Biodiversity Informatics Outlook: Delivering biodiver- sity knowledge in the information age.URL: https://doi.org/10.15468/6jxa-yb44
- Connecting data and expertise: a new alliance for biodiversity knowledge.Biodiversity Data Journal7https://doi.org/10.3897/bdj.7.e33679
- Data-intensive Science: A New Paradigm for Biodiversity Studies.BioScience59(7):613‑620. https://doi.org/10.1525/bio.2009.59.7.12
- Towards a unified paradigm for sequence-based identification of fungi.Molecular Ecology22(21):5271‑5277. https://doi.org/10.1111/mec.12481
- DiSSCo design study summary. URL: https://tinyurl.com/DiSSCo-Design-pdf
- FAIR Data and Services in Biodiversity Science and Geoscience.Data Intelligence2:122‑130. https://doi.org/10.1162/dint_a_00034
- Earth BioGenome Project: Sequencing life for the future of life.Proceedings of the National Academy of Sciences115(17):4325‑4333. https://doi.org/10.1073/pnas.1720115115
- Publishing the research process.Research Ideas and Outcomes1https://doi.org/10.3897/rio.1.e7547
- neXtA5: accelerating annotation of articles via automated approaches in neXtProt.Database2016https://doi.org/10.1093/database/baw098
- Triage by ranking to support the curation of protein interactions.Database2017https://doi.org/10.1093/database/bax040
- Reinventing Discovery: The New Era of Networked Science.Princeton University Press,Princeton, N.J.. [ISBN978-0-691-14890-8] https://doi.org/10.1515/9780691202853-004
- The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications.Nucleic Acids Research. https://doi.org/10.15468/6jxa-yb44
- Towards a biodiversity knowledge graph.Research Ideas and Outcomes2https://doi.org/10.3897/rio.2.e8767
- DNA barcoding and taxonomy: dark taxa and dark texts.Philosophical Transactions of the Royal Society B: Biological Sciences371(1702). https://doi.org/10.1098/rstb.2015.0334
- Ozymandias: a biodiversity knowledge graph.PeerJ7https://doi.org/10.7717/peerj.6739
- Semantic tagging of and semantic enhancements to systematics papers: ZooKeys working examples.ZooKeys50:1‑16. https://doi.org/10.3897/zookeys.50.538
- ARPHA-BioDiv: A toolbox for scholarly publication and dissemination of biodiversity data based on the ARPHA Publishing Platform.Research Ideas and Outcomes3https://doi.org/10.3897/rio.3.e13088
- OpenBiodiv: A Knowledge Graph for Literature-Extracted Linked Open Data in Biodiversity Science.Publications7(2). https://doi.org/10.3390/publications7020038
- Time to model all life on Earth.Nature493(7432):295‑297. https://doi.org/10.1038/493295a
- A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System.PLoS ONE8(7). https://doi.org/10.1371/journal.pone.0066213
- Making the Most of Your H2020 Project. Boosting the impact of your project through effective communication, dissemination and exploitation..European IPR Helpdesk. URL: https:// www.iprhelpdesk.eu/sites/default/files/EU-IPR-Brochure-Boosting-Impact-C-D-E_0.pdf
- OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system.Journal of Biomedical Semantics9(1). https://doi.org/10.1186/s13326-017-0174-5
- Next-generation sequencing technologies for environmental DNA research.Molecular Ecology21(8):1794‑1805. https://doi.org/10.1111/j.1365-294x.2012.05538.x
- Beyond dead trees: integrating the scientific process in the Biodiversity Data Journal.Biodiversity Data Journal1https://doi.org/10.3897/bdj.1.e995
- Eupolybothrus cavernicolus Komerički & Stoev sp. n. (Chilopoda: Lithobiomorpha: Lithobiidae): the first eukaryotic species description combining transcriptomic, DNA barcoding and micro-CT imaging data.Biodiversity Data Journal1https://doi.org/10.3897/bdj.1.e1013
- The metagenomic data life-cycle: standards and best practices.GigaScience6(8). https://doi.org/10.1093/gigascience/gix047
- Improving average ranking precision in user searches for biomedical research datasets.Database2017https://doi.org/10.1093/database/bax083
- Enriched biodiversity data as a resource and service.Biodiversity Data Journal 2https://doi.org/10.3897/BDJ.2.e11
Supplementary materials
Recommendation 3: Harmonise and integrate a vision for convergent operation of RIs and e-infrastructures in Europe to ensure cost-effective service provision to the user communities. https://www.esfri.eu/sites/default/files/ ESFRI_SCRIPTA_SINGLE_PAGE_19102017_0.pdf
See the Group of Senior Officials on Global Research Infrastructures Progress Report 2015. https://www.bmbf.de/ files/151109_G7_Broschere.pdf