|
Research Ideas and Outcomes :
Research Idea
|
|
Corresponding author: Michael Greeff (greeffm@ethz.ch)
Academic editor: Editorial Secretary
Received: 10 Dec 2021 | Accepted: 25 Jan 2022 | Published: 01 Mar 2022
© 2022 Michael Greeff, Max Caspers, Vincent Kalkman, Luc Willemse, Barry Sunderland, Olaf Bánki, Laurens Hogeweg
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Greeff M, Caspers M, Kalkman V, Willemse L, Sunderland BD, Bánki O, Hogeweg L (2022) Sharing taxonomic expertise between natural history collections using image recognition. Research Ideas and Outcomes 8: e79187. https://doi.org/10.3897/rio.8.e79187
|
|
Abstract
Natural history collections play a vital role in biodiversity research and conservation by providing a window to the past. The usefulness of the vast amount of historical data depends on their quality, with correct taxonomic identifications being the most critical. The identification of many of the objects of natural history collections, however, is wanting, doubtful or outdated. Providing correct identifications is difficult given the sheer number of objects and the scarcity of expertise. Here we outline the construction of an ecosystem for the collaborative development and exchange of image recognition algorithms designed to support the identification of objects. Such an ecosystem will facilitate sharing taxonomic expertise among institutions by offering image datasets that are correctly identified by their in-house taxonomic experts. Together with openly accessible machine learning algorithms and easy to use workbenches, this will allow other institutes to train image recognition algorithms and thereby compensate for the lacking expertise.
Keywords
Digitization, image recognition, taxonomic expertise, herbaria, natural history collections
Overview and background
Worldwide there are thousands of repositories housing natural history collections (
Taxonomic identifications guarantee collection accessibility
To make full use of natural history collections, both their physical and digital visibility and accessibility are crucial. Physical accessibility is linked to the degree of management applied to collections (for an overview of collection management levels, see
In most repositories, collections cover large parts of the biodiversity often from all bioregions of the world. The larger the taxonomic and geographic scope of a collection the more taxonomic expertise and working time is required for its identification. For quite a while, however, there has been a trend for taxonomy to receive less and less attention in the curricula of universities, and positions in public institutions incorporating traditional taxonomy were filled with staff with no or only little taxonomic expertise. This trend, coined the taxonomic impediment (
Likewise, the degree of digital data capturing not only depends on capacity and funding but to a large degree also on the systematic organization of a collection, which can only be done if specimens have proper taxonomic identifications. In line with this, the Minimum Standard for Digital Specimens (MIDS), which was developed for the Distributed System of Scientific Collections DiSSCo (www.dissco.eu;
Image recognition to the rescue
As discussed in the previous sections, taxonomic knowledge is distributed very unevenly and resources for taxonomic work are scarce. For many years, there have been calls for collaboration between taxonomists and specialists in artificial intelligence, machine learning, and pattern recognition to develop automated systems capable of conducting high-throughput identification of biological specimens (
Especially in the context of national and international digitization initiatives such as DiSSCo, the Integrated Digitized Biocollections iDigBio (www.idigbio.org,
Although machine learning solutions are getting ever more powerful and capable of identifying diverse objects, a single universal machine learning model for all known biological taxa is still technically challenging and costly. As a reasonable solution for the time being, collection staff therefore need machine learning tools focusing on subsets of biodiversity such as organisms from limited geographical areas and/or limited taxonomic groups. For instance, machine learning models have been developed for British ground beetle species (
Automated identifications are transparent and reproducible
Recent studies proved that machine identifications have become almost as accurate as identifications done by human experts in quite a few groups (in benthic macroinvertebrates (
In contrast to identifications done by human experts, machine identifications not only deliver taxonomic names, but also metadata about the probability of the determination, the range of taxa considered, the version of the application, and other parameters. Machine determinations therefore are quantifiable, transparent, and reproducible by anyone (the data management techniques involved fall under the term provenance which help reproduce, trace, assess, understand, and explain models and how they were constructed). As natural history collections data are increasingly used in statistical modeling of environmental changes and large datasets are assembled from different repositories, transparent identifications become ever more important (
Objectives
An automated image recognition ecosystem
The authors envision the establishment of a machine learning ecosystem for natural history collections which allows the sharing of existing models, image datasets and know-how between institutions and collection personnel. An avant-garde of a few experienced institutions shall develop the necessary core modules in machine learning, which can easily be re-trained by other institutions to serve their individual needs. This ecosystem should rest on four pillars:
- a central library of machine learning algorithms and associated applications (e.g. mobile apps)
- a central library of available expert validated training datasets, be it the images themselves or simply the information where to find these images
- a digital workbench that allows even inexperienced users to customize existing machine learning solutions to their individual needs
- a user forum for the discussion of problems and the coordination of next steps, for the evaluation, testing and implementation of novel technologies, etc.
Deep learning
Feature extractor. Deep learning models (
Classifier. The feature extractor does not relate the resulting categories to explicit human concepts such as animals, plants, or cars. For this, the machine learning model relies on a classifier network, which associates the output of the feature extractor with names and concepts (i.e., "classes"). In the natural history context, for instance, the classifier would associate certain features with a family of plants, a species of beetle etc. Classifiers can be easily (re)trained, with regard to time, computing power and experience of the user (e.g., see
Algorithms. Machine learning models make predictions and are trained in a particular way and with a particular dataset as described above. Using models in practice often involves additional functionality. The complete process from image(s) to identifications can generally be described as an algorithm. Besides the models themselves, algorithms contain pre- and post-processing functionality that cannot be easily fitted into the model formalism of a feature extractor and a classifier. An example of pre-processing is explicitly localizing the organism in the picture before identification. Examples of post-processing are combining multiple predictions into one and combining image recognition models with species distribution models.
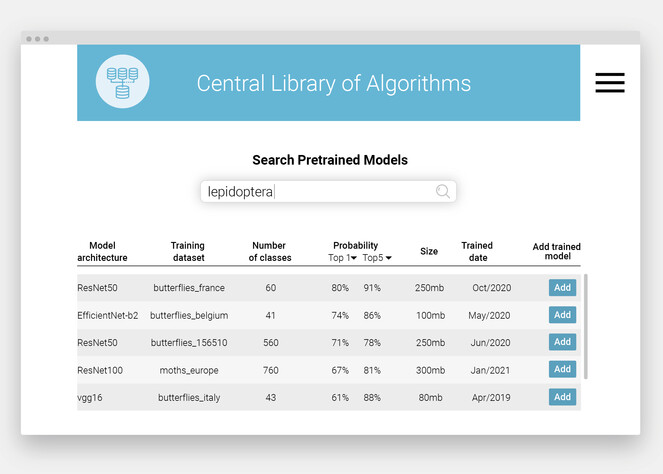
Central Library of Algorithms. To facilitate the exchange of these models and algorithms, the authors suggest setting up a Central Library of Algorithms (Fig.
Central Library of Datasets
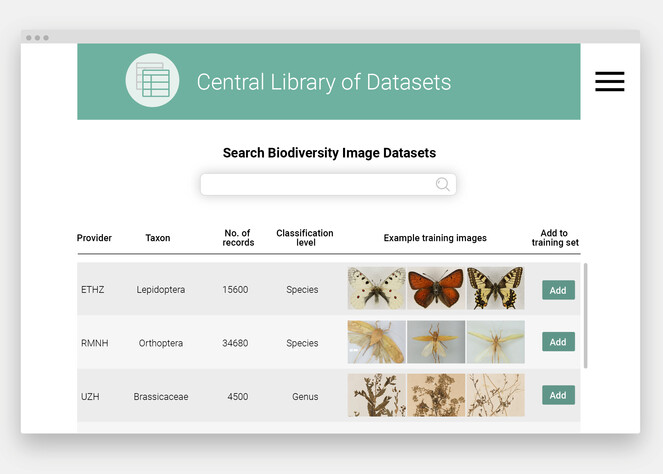
Further sharing of taxonomic knowledge would be provided through a Central Library of Datasets. This library would be a system to access a collection of public image datasets for images that are suitable for supporting large-scale centralized training of feature extractors and local training of classifiers at the individual institutions (Fig.
In the Central Library of Datasets, natural history collection staff will find correctly identified images of their target organisms and download the data for training of an individually customized classifier (photos: Lepidoptera by Entomological Collection of ETH Zürich; Orthoptera by Naturalis Biodiversity Center; Brassicaceae by United Herbaria Z+ZT, ZT-00164967, ZT-00167494, ZT-00171530, CC BY-SA 4.0). The current figure shows a mock-up.
The uploaded images shall be collected in a dataset, in this context defined as a fixed curated list of images with additional metadata such as the name of the taxon, geographic coordinates and information on the probability of the identification. The Central Library of Datasets will reference existing public datasets such as GBIF and/or iDigBio. Over time, this can encourage collection staff and collection users to generate and publish their own datasets on public portals, possibly remedying biases and shortcomings in existing datasets (this could be done as 'data papers', see
Digital workbench
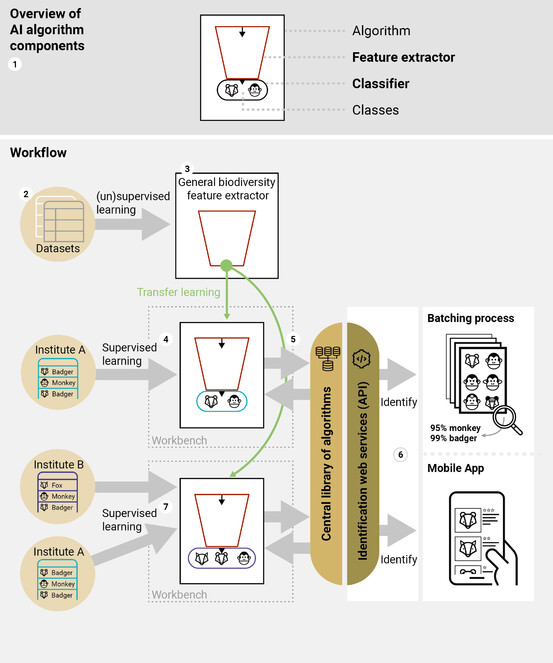
Retraining an existing model to a new group of organisms is easy – for IT specialists. The average collection manager would most likely struggle with the necessary procedures. The authors therefore propose the establishment of a digital workbench for machine learning (e.g., Google AutoML, Microsoft Azure), which allows non-experts to curate datasets (e.g., completing taxonomic or geographic information) and retrain existing models for their individual purposes. Ideally, the workbench should have a graphical user interface. Users could import existing feature extractors and further algorithms from the Central Library of Algorithms, and training data from the Central Library of Datasets (Fig.
Sharing of taxonomic knowledge between institutes. (1) Each algorithm contains two basic components: the feature extractor and the classifier. (2) The Central Library of Datasets allows the user to browse through all available images of collection objects; (3) based on all available images, a regularly updated central feature extractor is created and published; (4) custom made algorithms can relatively easily be created by building a classifier based on a selection of taxa from the central library and combining this with the central feature extractor; (5) newly created algorithms together with their metadata (probability & information on content) are published through a web service in the Central Library of Algorithms (6) and can be used through the Identification web services (API) either for batch processing of images or through a mobile app. Models can be easily extended by other institutions by combining data sources (7).
User forum
Critical readers might consider this vision too idealistic. And it is true, for everything to work properly, many prerequisites just need to be right: a feature extractor needs to be available, appropriate images need to exist, the workbench and the applications need to work flawlessly. The authors therefore propose a further measure: the establishment of a user forum. On this forum, users can post their wishes, discuss shortcomings, and interact with more experienced institutions and providers of machine learning solutions. The user forum should thus serve as a marketplace where collection managers search for technological expertise and assistance and in return offer image datasets and taxonomic expertise. As a result, this user forum should guarantee that over time well identified image datasets and machine learning models become available for most groups of organisms, as well those that have been neglected so far. In addition, this will be the place to discuss and find strategies for shortcomings of the AI solutions related to inherent collection biases, be they geographical, cultural, taxonomical or other.
Use Cases
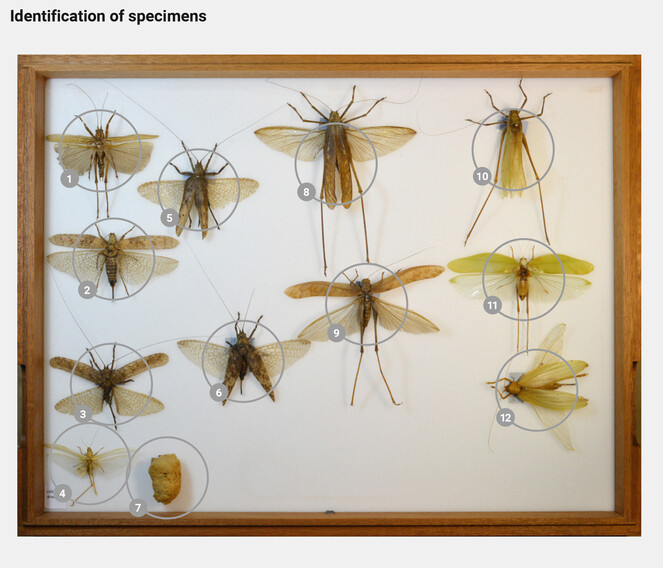
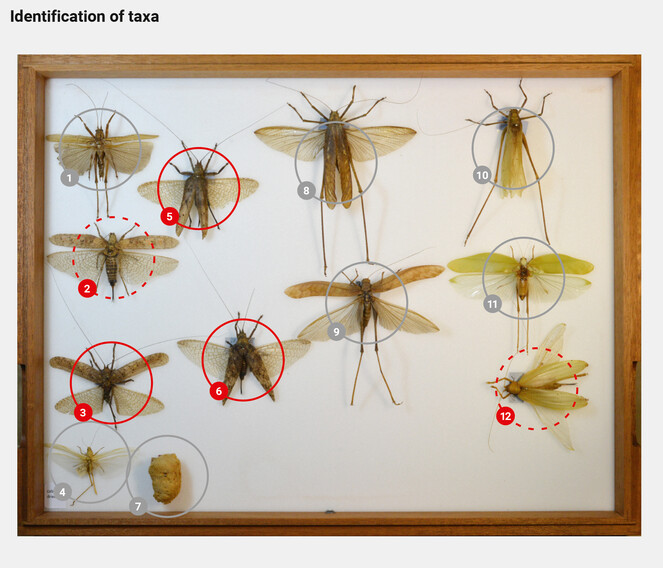
Accessing unsorted collection holdings. Most collections accumulate considerable holdings of biological specimens which remain unidentified due to a lack of time or in-house taxonomic expertise. These specimens may be stored as singletons or as groups in boxes, either preliminarily sorted by higher taxonomic groupings (order, family) or by geographic region, or they may be completely mixed. In recent years, especially larger institutions have therefore started to database their holdings at the storage unit level (i.e., by the units in which specimens are stored, like drawers, jars, or boxes). In insect collections, for instance, whole drawers are being imaged and published online to be browsed through by the entomological community (
Machine learning applications would then recognize the taxonomic identity of each specimen (Table
Automated recognition applications identify the specimens to lower taxonomic levels and inform about the probability of the identifications.
|
Drawer number |
Specimen number |
Family |
Subfamily |
Probability |
|
BE.2286032 |
1 |
Tettigoniidae |
Conocephalinae |
95% |
|
BE.2286032 |
2 |
Tettigoniidae |
Pseudophyllinae |
85% |
|
BE.2286032 |
3 |
Tettigoniidae |
Pseudophyllinae |
95% |
|
... |
... |
... |
... |
... |
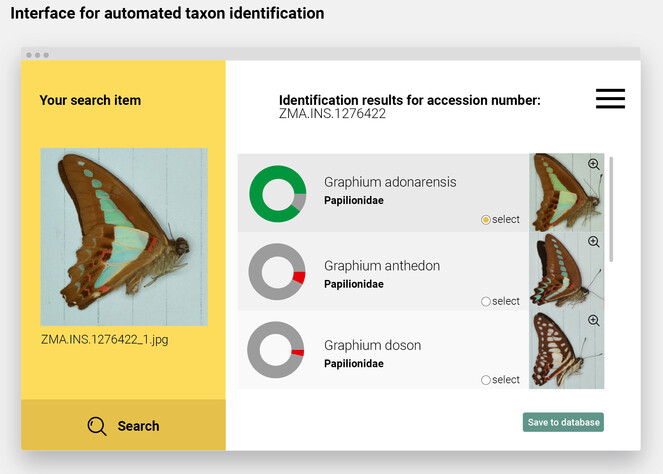
Transparent identifications in mass digitization. Bringing down costs and time spent per treated item is of paramount importance when digitizing natural history collections (
Mock-up of an interface for automated taxon identification. Naturalis holds over 500.000 specimens of unmounted, unsorted and often unidentified, papered butterflies and moths that were collected mostly in Europe and Asia over the past 200 years. In early 2016, Naturalis embarked on a 10-year-project to digitally identify all these specimens with the help of dedicated volunteers (
Challenges ahead
A decade ago, the idea of using image recognition to share taxonomic knowledge between natural history collections would have seemed far-fetched. From a technical point of view this is no longer the case as is demonstrated by widely used field apps like iNaturalist, ObsIdentify (
Algorithms. Even though most challenges ahead are organizational, machine learning still harbors some technical challenges of its own (e.g.,
Standardization. One organizational endeavor is to further standardize and accelerate the digitization of natural history collections, ensuring that the images and metadata can be readily applied for image recognition. This applies to both taxonomical and geographical annotations. Even when no larger infrastructure as envisioned in this paper is built, this step is worthwhile and should be addressed by or in close collaboration with TDWG (
Infrastructure. Another challenge is the ownership and responsibility for the proposed ecosystem. Initially, one or several larger natural history institutions will need to build a large-scale digital infrastructure to allow for the generation, exchange, and application of image recognition models, as well as to provide a platform for a community to engage with one another. The different modules of the infrastructure can be developed by different parties. In addition, the different modules could be a combination of the repurposing of existing infrastructure components and tools and newly developed ones. Recently, a landscape and gap analysis on the automated services, tools, and workflows for extracting information from images of natural history specimens and their labels was performed (
Once built, the viability of the machine learning ecosystem for collections depends on the level of contribution from its participants. Collection managers and curators would need to actively focus their capacities at collaborating with experts to identify and digitize collections, resulting in taxonomically validated and properly annotated images. Once shared, they can be used to (re)train image recognition models and benefit the entire community. Especially in the initial phase this will require a level of altruism, as contributing will take time and resources while the benefits will only become clear after a few years. The concept of give and take requires momentum and should be stimulated by the collections maintaining the infrastructure, ideally utilizing already existing cross-national collaborations for mobilizing collections and knowledge. Parallels of such a community-driven approach can be found in the Barcode of Life project (www.barcodinglife.org), which allows the exchange of DNA-barcodes between institutes, or OpenML (
Acknowledgements
We are especially grateful to Rod Eastwood and Samuel Glauser for their discussion of and feedback on the current text.
References
- Human experts vs. machines in taxa recognition.Signal Processing: Image Communication87:115917. https://doi.org/10.1016/j.image.2020.115917
- The Global Museum: natural history collections and the future of evolutionary science and public education.PeerJ8:e8225. https://doi.org/10.7717/peerj.8225
- Catalogue of Life Plus: innovating the CoL systems as a foundation for a clearinghouse for names and taxonomy.Biodiversity Information Science and Standards2https://doi.org/10.3897/biss.2.26922
- Towards Open Set Deep Networks.2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR)https://doi.org/10.1109/cvpr.2016.173
- No specimen left behind: industrial scale digitization of natural history collections.ZooKeys209:133‑146. https://doi.org/10.3897/zookeys.209.3178
- What is an expert? A systems perspective on expertise.Ecology and Evolution4(3):231‑242. https://doi.org/10.1002/ece3.926
- Unsupervised learning of visual features by contrasting cluster assignments.arXiv preprint(arXiv:2006.09882). URL: https://arxiv.org/pdf/2006.09882.pdf
- Butterflies in bags: permanent storage of Lepidoptera in glassine envelopes.Nota Lepidopterologica42(1):1‑16. https://doi.org/10.3897/nl.42.28654
- The data paper: a mechanism to incentivize data publishing in biodiversity science.BMC Bioinformatics12https://doi.org/10.1186/1471-2105-12-s15-s2
- Big self-supervised models are strong semi-supervised learners.arXiv preprint(arXiv:2006.10029). URL: https://arxiv.org/pdf/2006.10029.pdf
- Biodiversity data should be published, cited, and peer reviewed.Trends in Ecology & Evolution28(8):454‑461. https://doi.org/10.1016/j.tree.2013.05.002
- Do experts make mistakes? A comparison of human and machine identification of dinoflagellates.Marine Ecology Progress Series247:17‑25. https://doi.org/10.3354/meps247017
- Hierarchical Image Classification using Entailment Cone Embeddings.2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW)https://doi.org/10.1109/cvprw50498.2020.00426
- Research and Societal Benefits of the Global Biodiversity Information Facility.BioScience54(6):485‑486. https://doi.org/10.1641/0006-3568(2004)054[0486:rasbot]2.0.co;2
- How reliable are species identifications in biodiversity big data? Evaluating the records of a neotropical fish family in online repositories.Systematics and Biodiversity18(2):181‑191. https://doi.org/10.1080/14772000.2020.1730473
- SwissCollNet – A National Initiative for Natural History Collections in Switzerland.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37188
- Automated species identification: why not?Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences359(1444):655‑667. https://doi.org/10.1098/rstb.2003.1442
- Recent Advances in Open Set Recognition: A Survey.IEEE Transactions on Pattern Analysis and Machine Intelligence43(10):3614‑3631. https://doi.org/10.1109/tpami.2020.2981604
- Visual-based plant species identification from crowdsourced data.Proceedings of the 19th ACM international conference on Multimedia - MM '11813‑814. https://doi.org/10.1145/2072298.2072472
- Multi-organ plant identification.Proceedings of the 1st ACM international workshop on Multimedia analysis for ecological data - MAED '12https://doi.org/10.1145/2390832.2390843
- Widespread mistaken identity in tropical plant collections.Current Biology25(22). https://doi.org/10.1016/j.cub.2015.10.002
- Deep learning for visual understanding: A review.Neurocomputing187:27‑48. https://doi.org/10.1016/j.neucom.2015.09.116
- Species‐level image classification with convolutional neural network enables insect identification from habitus images.Ecology and Evolution10(2):737‑747. https://doi.org/10.1002/ece3.5921
- Provisional Data Management Plan for DiSSCo infrastructure.ICEDIG Deliverable D6.6https://doi.org/10.5281/zenodo.3532937
- Conceptual design blueprint for the DiSSCo digitization infrastructure - DELIVERABLE D8.1.Research Ideas and Outcomes6https://doi.org/10.3897/rio.6.e54280
- Why ReLU Networks Yield High-Confidence Predictions Far Away From the Training Data and How to Mitigate the Problem.2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR):41‑50. https://doi.org/10.1109/cvpr.2019.00013
- Deep Residual Learning for Image Recognition.2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR):770‑778. https://doi.org/10.1109/cvpr.2016.90
- The taxonomic impediment and the convention on Biodiversity.Association of Systematics Collections Newsletter24:61‑62.
- Advancing the Catalogue of the World’s Natural History Collections.Biodiversity Information Science and Standards4https://doi.org/10.3897/biss.4.59324
- Declines in the numbers of amateur and professional taxonomists: implications for conservation.Animal Conservation5(3):245‑249. https://doi.org/10.1017/s1367943002002299
- Deep learning and computer vision will transform entomology.Proceedings of the National Academy of Sciences118(2). https://doi.org/10.1073/pnas.2002545117
- Aleatoric and epistemic uncertainty in machine learning: an introduction to concepts and methods.Machine Learning110(3):457‑506. https://doi.org/10.1007/s10994-021-05946-3
- Overview of LifeCLEF 2020: A System-Oriented Evaluation of Automated Species Identification and Species Distribution Prediction.Lecture Notes in Computer Science342‑363. https://doi.org/10.1007/978-3-030-58219-7_23
- An algorithm competition for automatic species identification from herbarium specimens.Applications in plant sciences8(6):e11365. https://doi.org/10.1002/aps3.11365
- Time to automate identification.Nature467(7312):154‑155. https://doi.org/10.1038/467154a
- Whole-drawer imaging for digital management and curation of a large entomological collection.ZooKeys209:147‑163. https://doi.org/10.3897/zookeys.209.3169
- A Computational- and Storage-Cloud for Integration of Biodiversity Collections.Proceedings of the IEEE 9th International Conference on e-Science:78‑87. https://doi.org/10.1109/escience.2013.48
- Where’s the management in collections management?International Symposium and First World Congress on the preservation and conservation of Natural History Collections3:309‑338.
- Biological collections for understanding biodiversity in the Anthropocene.Philosophical Transactions of the Royal Society B: Biological Sciences374:20170386. https://doi.org/10.1098/rstb.2017.0386
- Application of deep learning in aquatic bioassessment: Towards automated identification of non-biting midges.Science of The Total Environment711:135160. https://doi.org/10.1016/j.scitotenv.2019.135160
- Machine Learning.McGraw-Hill Education,414pp.
- Discovery and publishing of primary biodiversity data associated with multimedia resources: The Audubon Core strategies and approaches.Biodiversity Informatics8(2):185‑197. https://doi.org/10.17161/bi.v8i2.4117
- Biological Collections: Ensuring Critical Research and Education for the 21st Century.The National Academies Press,Washington,245pp. [ISBNISBN 978-0-309-49853-1] https://doi.org/10.17226/25592
- Deep neural networks are easily fooled: High confidence predictions for unrecognizable images.Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR):427‑436. https://doi.org/10.1109/cvpr.2015.7298640
- ‘From Pilot to production’: Large Scale Digitisation project at Naturalis Biodiversity Center.ZooKeys209:87‑92. https://doi.org/10.3897/zookeys.209.3609
- Museum specimens find new life online.New York Times.
- The research data alliance: Implementing the technology, practice and connections of a data infrastructure.Bulletin of the American Society for Information Science and Technology39(6):33‑36. https://doi.org/10.1002/bult.2013.1720390611
- White paper on the alignment and interoperability between the Distributed System of Scientific Collections (DiSSCo) and EU infrastructures - The case of the European Environment Agency (EEA).Research Ideas and Outcomes6https://doi.org/10.3897/rio.6.e62361
- Supporting citizen scientists with automatic species identification using deep learning image recognition models.Biodiversity Information Science and Standards2https://doi.org/10.3897/biss.2.25268
- Applications of deep convolutional neural networks to digitized natural history collections.Biodiversity data journal5:e21139. https://doi.org/10.3897/BDJ.5.e21139
- Rationale and Value of Natural History Collections Digitisation.Biodiversity Informatics7(2):77‑80. https://doi.org/10.17161/bi.v7i2.3994
- Very deep convolutional networks for large-scale image recognition.arXiv preprint(arXiv:1409.1556). URL: https://arxiv.org/pdf/1409.1556.pdf(2014.pdf
- Bayesian Methods for Intelligent Task Assignment in Crowdsourcing Systems.Decision Making: Uncertainty, Imperfection, Deliberation and Scalability1‑32. https://doi.org/10.1007/978-3-319-15144-1_1
- Workflow provenance in the lifecycle of scientific machine learning.Concurrency and Computation: Practice and Experiencehttps://doi.org/10.1002/cpe.6544
- The Value of Museum Collections for Research and Society.BioScience54(1):66‑74. https://doi.org/10.1641/0006-3568(2004)054[0066:tvomcf]2.0.co;2
- BioDex: Tales from an Adventure into App Development.ETH Library Lab BlogURL: https://www.librarylab.ethz.ch/tales-from-an-adventure-into-app-development/
- Going deeper with convolutions.Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR):1‑9. https://doi.org/10.1109/cvpr.2015.7298594
- Rethinking the Inception Architecture for Computer Vision.Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR):2818‑2826. https://doi.org/10.1109/cvpr.2016.308
- Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning?IEEE Transactions on Medical Imaging35(5):1299‑1312. https://doi.org/10.1109/tmi.2016.2535302
- High-performance digitization of natural history collections: Automated imaging lines for herbarium and insect specimens.Taxon63(6):1307‑1313. https://doi.org/10.12705/636.13
- Taxonomic bias in biodiversity data and societal preferences.Scientific Reports7(1):1‑4. https://doi.org/10.1038/s41598-017-09084-6
- iNaturalist as an engaging tool for identifying organisms in outdoor activities.Journal of Biological Education1‑11. https://doi.org/10.1080/00219266.2020.1739114
- Automated Taxonomic Identification of Insects with Expert-Level Accuracy Using Effective Feature Transfer from Convolutional Networks.Systematic Biology68(6):876‑895. https://doi.org/10.1093/sysbio/syz014
- Automated image-based taxon identification using deep learning and citizen-science contributions. Doctoral Dissertation.Department of Zoology, Stockholm University
- OpenML.ACM SIGKDD Explorations Newsletter15(2):49‑60. https://doi.org/10.1145/2641190.2641198
- Automated plant species identification—Trends and future directions.PLOS Computational Biology14(4). https://doi.org/10.1371/journal.pcbi.1005993
- Landscape Analysis for the Specimen Data Refinery.Research Ideas and Outcomes6https://doi.org/10.3897/rio.6.e57602
- Darwin Core: An Evolving Community-Developed Biodiversity Data Standard.PLoS ONE7(1). https://doi.org/10.1371/journal.pone.0029715
- Join the Dots: Adding collection assessment to collection descriptions.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37200
- How transferable are features in deep neural networks?arXiv preprint(arXiv:1411.1792). URL: https://arxiv.org/pdf/1411.1792.pdf