|
Research Ideas and Outcomes :
Grant Proposal
|
|
Corresponding author: W. Daniel Kissling (wdkissling@gmail.com)
Received: 08 Jul 2017 | Published: 16 Jul 2017
© 2017 W. Daniel Kissling, Arie Seijmonsbergen, Ruud Foppen, Willem Bouten
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Kissling WD, Seijmonsbergen AC, Foppen RPB, Bouten W (2017) eEcoLiDAR, eScience infrastructure for ecological applications of LiDAR point clouds: reconstructing the 3D ecosystem structure for animals at regional to continental scales. Research Ideas and Outcomes 3: e14939. https://doi.org/10.3897/rio.3.e14939
|
|
Abstract
The lack of high-resolution measurements of 3D ecosystem structure across broad spatial extents impedes major advancements in animal ecology and biodiversity science. We aim to fill this gap by using Light Detection and Ranging (LiDAR) technology to characterize the vertical and horizontal complexity of vegetation and landscapes at high resolution across regional to continental scales. The newly LiDAR-derived 3D ecosystem structures will be applied in species distribution models for breeding birds in forests and marshlands, for insect pollinators in agricultural landscapes, and songbirds at stopover sites during migration. This will allow novel insights into the hierarchical structure of animal-habitat associations, into why animal populations decline, and how they respond to habitat fragmentation and ongoing land use change. The processing of these massive amounts of LiDAR point cloud data will be achieved by developing a generic interactive eScience environment with multi-scale object-based image analysis (OBIA) and interpretation of LiDAR point clouds, including data storage, scalable computing, tools for machine learning and visualisation (feature selection, annotation/segmentation, object classification, and evaluation), and a PostGIS spatial database. The classified objects will include trees, forests, vegetation strata, edges, bushes, hedges, reedbeds etc. with their related metrics, attributes and summary statistics (e.g. vegetation openness, height, density, vertical biomass distribution etc.). The newly developed eScience tools and data will be available to other disciplines and applications in ecology and the Earth sciences, thereby achieving high impact. The project will foster new multi-disciplinary collaborations between ecologists and eScientists and contribute to training a new generation of geo-ecologists.
Keywords
3D vegetation & landscape structure, LiDAR, Object Based Image Analysis (OBIA), eScience research infrastructure, ecological modelling, quantitative biodiversity science
List of participants
Core research team
- Zsófia Koma (PhD student, University of Amsterdam, Amsterdam, The Netherlands)
- Christiaan Meijer (eScience Research Engineer, Netherlands eScience Center, Amsterdam, The Netherlands)
- W. Daniel Kissling (Associate Professor, University of Amsterdam, Amsterdam The Netherlands)
- Arie C. Seijmonsbergen (Assistant Professor, University of Amsterdam, Amsterdam, The Netherlands)
- Willem Bouten (Professor, University of Amsterdam, Amsterdam, The Netherlands)
- Ruud P. B. Foppen (Professor, Radboud University, Nijmegen, The Netherlands)
Management support
- Elena Ranguelova (eScience coordinator, Netherlands eScience Center, Amsterdam, The Netherlands)
Associated Master students
- Tristan Bakx (MSc student, University of Amsterdam, Amsterdam, The Netherlands)
- Jim Groot (MSc student, University of Amsterdam, Amsterdam, The Netherlands)
- Berend Wijers (MSc student, University of Amsterdam, Amsterdam, The Netherlands)
- Peter Haacke (MSc student, University of Amsterdam, Amsterdam, The Netherlands)
- Chris Lucas (MSc student, University of Amsterdam, Amsterdam, The Netherlands)
Associated researchers
- Adriaan M. Dokter (Postdoc, Cornell Lab of Ornithology, Cornell University, USA)
- Jesus Aguirre-Gutiérrez (Postdoc, Environmental Change Institute, University of Oxford, UK)
- Henk Sierdsema (Researcher, Sovon Vogelonderzoek Nederland, Nijmegen, The Netherlands)
- Marco Heurich (Deputy head, Department of Conservation and Research, Bavarian Forest National Park, Grafenau, Germany)
Advisory board
- Jacobus C. Biesmeijer (Professor, Naturalis Biodiversity Center, Leiden, The Netherlands)
- Peter J.M. van Oosterom (Professor, Delft University of Technology, Delft, The Netherlands)
- Andrew K. Skidmore (Professor, University of Twente, Enschede, The Netherlands)
- Maurice Bouwhuis (Computational scientist, SURFsara, Amsterdam, The Netherlands)
- Hans van Gasteren (Head of Nature Bureau, Royal Netherlands Air Force, Gouda, The Netherlands)
- Niels S. Anders (Researcher, Geodan, Amsterdam, The Netherlands)
Science: background, research questions, approach, and innovation
Humans have a tremendous impact on the natural environment. For instance, human-modified landscapes are now dominating our planet and the conversion, degradation and loss of habitat leads to species extinctions and severely affects the distribution of species and ecosystems and the services they provide to humanity
A major bottleneck for predictive biodiversity modelling is the current lack of high-resolution (i.e. fine-scale) measurements of habitat structures and 3D characteristics of vegetation across regions and continents (
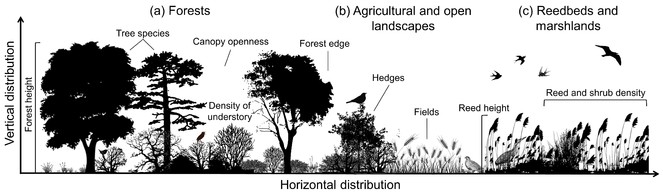
The vertical and horizontal distribution of plants influences habitat structure and 3D characteristics of vegetation for animals. Illustrated are examples for (a) forests, (b) agricultural and open landscapes, and (c) reedbeds and marshlands. The height, openness and density of vegetation as well as specific habitat features (e.g. tree species, hedges etc.) are key aspects of animal habitat and space use.
An exciting opportunity to improve predictive biodiversity modelling is the increasing availability of high-resolution remote sensing (RS) data on ecosystem structures derived from Light Detection and Ranging (LiDAR). LiDAR data enable us to fill the existing data gap by providing fine-scale habitat information across large spatial extents (
The aim of this project is to use LiDAR technology to quantify fine-scale 3D ecosystem structures across broad spatial extents. Across Europe, we will focus on (1) ground-nesting breeding birds in forests (e.g. Wren, Wood Warbler, Common Nightingale etc.) for which the 3D forest structure (e.g. forest height, stem density, canopy openness, density of understory etc.) is of key relevance (Fig.
We will expose the developed eScience infrastructure to two other ongoing research projects, thus increasing the impact, generating user feed-back, and identifying bottlenecks for wider applicability. Postdoc J. Aguirre-Gutiérrez focuses on the impact of land-use change on the distribution and loss of NW-European pollinators. This includes insects such as bees, butterflies, and hoverflies. Predictive modelling is currently restricted to coarse-scale 2D habitat characterization (
The proposed project will enable scientific breakthroughs in predicting animal populations and species distributions at much finer resolution and higher accuracy than ever has been previously possible. This will strongly push the frontiers of ecology, biogeography and conservation by providing new data and novel insights into the distribution of biodiversity and ecosystems. The availability of LiDAR-derived 3D ecosystem structures across broad spatial extents will have a major impact, maybe comparable to the influence of the WorldClim dataset (
eScience: technologies, methods, and expected impact on the research
We are witnessing changes in remote sensing (RS) from grid cell-based approaches to object-based approaches (
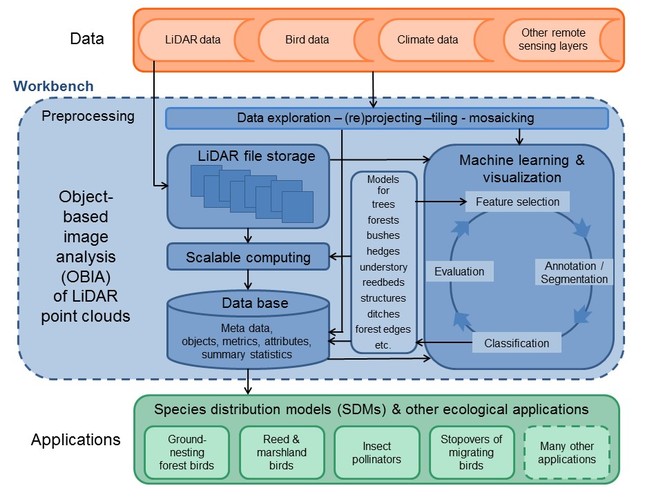
The methodological and technological aim of the proposed project is to develop a workbench that supports the workflow for handling, storage, and interactive object-based image analysis (OBIA) of massive amounts of LiDAR point cloud data (Fig.
Generic workflow for object-based image analysis (OBIA) of LiDAR point clouds and proposed ecological applications. A workbench (blue) will be developed to handle the data storage, data exploration, and interactive OBIA of the massive LiDAR point clouds. Combined with datasets of bird distributions, climate, and other remote sensing layers (orange), the LiDAR data will be applied to several ecological case studies, e.g. by using species distribution modelling of birds and insect pollinators (green).
We already have many TeraBytes of LiDAR-data (NL, BE, AUT), we are in contact with some other countries (GB, DE), and many European countries have LiDAR data that are available for scientific research (e.g. ES, FIN, DK, SI). We will take care of the differences between data sets in terms of point cloud density and information type (e.g. full wave form, intensity, only first return, additional parallel sensors). Uniform global LiDAR data will become accessible when the GEDI sensor is installed on the International Space Station in 2018 (https://www.nasa.gov/content/goddard/new-nasa-probe-will-study-earth-s-forests-in-3-d/#.VzWjAr4T5fA). Besides LiDAR, we have access to bird distribution and abundance data, climate data, and other remote sensing layers such as Sentinel imagery (https://scihub.copernicus.eu), Landsat imagery (http://landsat.gsfc.nasa.gov), and SPOT vegetation products such as NDVI (http://www.vgt.vito.be/index.html). These will be needed for species distribution modelling. For ground truthing of LiDAR data and derived objects, we will use specific test areas in flat as well as mountainous regions (e.g. cultural landscapes in the Netherlands vs. steep slopes in the Alps) to assess the accuracy of the identification of trees, understory density, shrubs, hedges, marshland habitats etc. in different environments. We will then develop and test the workflow for supervised OBIA to capture the full variation of vegetation across Europe.
LAStools (https://rapidlasso.com/lastools/) are indispensable for efficient LiDAR processing (i.e. converting, tiling, filtering, and clipping the many TeraBytes of data). However, additional tools are needed for efficient and transparent object-based classification. We aim to combine OBIA with scalable computing in an interactive environment for data exploration, segmentation, classification and interpretation of LiDAR data. The following elements for developing the workflow are essential:
- Scalable storage. We will use existing file-based LAStools for storing LiDAR data. In addition, we will develop a PostgreSQL/GIS database for storing the metadata of the LiDAR and other RS data, and for storing classified objects with their attributes and summary statistics. It should be easy for a user to add new schemas and new objects to the data base. Objects may have different spatial scales (e.g. single trees in a forest, the forest as a whole, hedges or other linear structures, reedbeds etc.). Attributes of e.g. a tree may be tree metrics such as height, crown diameter, biomass, species identity etc.. The data base with objects will be made available to scientists (ecologists, environmental scientists, meteorologists) which we expect will increase the impact of our work enormously (see deliverables below).
- Tools for machine learning. Traditional tools for geospatial analyses are not yet ready for point clouds. eCognition (http://www.ecognition.com/) is a suite of commercial software for interactive OBIA (including point clouds), but capabilities are still very limited and classification algorithms are (partly) hidden and thus inappropriate for scientific innovation. The challenge in the proposed project is to make use of existing open source software such as the Point Cloud Library (
Aldoma et al. 2012 , http://pointclouds.org/), CloudCompare (http://www.danielgm.net/cc/), and Orfeo (https://www.orfeo-toolbox.org/) and to include machine learning; to closely monitor and make use of new developments; and to complement this by developing new software. From our previous research we have good experience with the WEKA software (Jakubowski et al. 2013 ,Frank et al. 2010 ) which covers various machine learning algorithms and data preprocessing tools. We therefore know that several existing machine learning algorithms can provide good results for pattern recognition as long as annotation, segmentation (see Box 1 for an example) and data features are optimally taken care of. The development of algorithms to calculate new features as well as feature selection have emerged as key steps. We will therefore mainly invest in tools for interactive feature selection and annotation/segmentation relevant for object classification (Anders et al. 2011 ). - Visualization. In the iterative process of feature selection, annotation/segmentation, and object classification and evaluation, the visualization is of utmost importance, especially when working on the improvement of methodologies. To be able to visually judge the quality of methodologies and to understand mismatches, we will adapt existing software such as Point Cloud Library (
Aldoma et al. 2012 , http://pointclouds.org/) and Potree (http://potree.org/wp/) for the visualization of merged objects and point clouds. - Computation. Once the models and methods for classification of objects (e.g. trees, bushes, hedges, reedbeds etc.; Fig.
2 ) have been developed, we aim to apply them to large areas (regions to continents). This will require to develop scalable computing solutions.
The eScience engineers will develop the proposed workbench (we envisage a combination of LAStools, Point Cloud Library, CloudCompare, Orfeo, QGIS, Potree, as well as newly developed tools) in close collaboration with the PhD student who will characterize the 3D ecosystem structures for breeding birds (see above), while the postdocs working on pollinators (
Box 1: Identifying trees in a forest
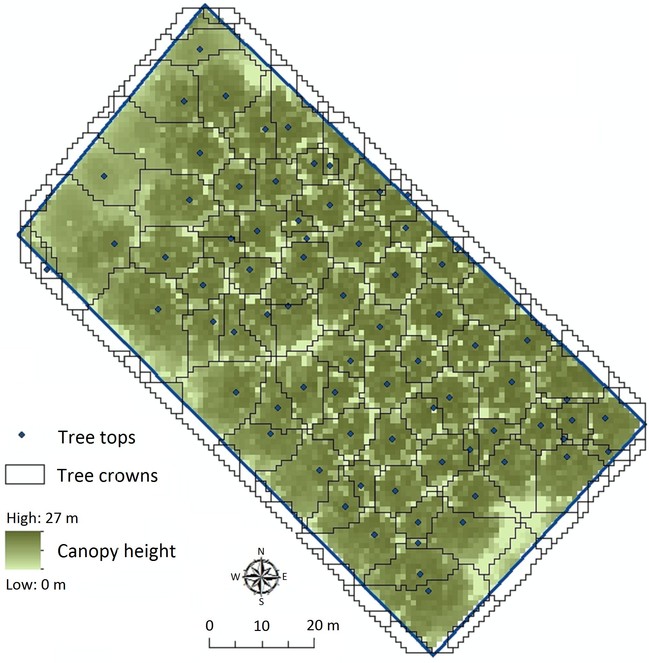
LiDAR data can be used to delineate individual trees in forests (
LiDAR returns were filtered from the point cloud, smoothed and rasterized to a 1m resolution Canopy Height Model (CHM). Locations of tree tops were determined using a local maximum filter and the minimum distance between trees. The CHM was then flipped with tree tops becoming sinks. An existing algorithm (
Our example shows how tree crowns and tree tops can be calculated from LiDAR data (Fig.
Re-use, sustainability, dissemination, and collaborations
The use of LiDAR point clouds is dramatically increasing. A generic challenge across many disciplines and applications is the storage and handling of massive amounts of data, the visualization, and the automated identification of objects (models) prior to the actual interpretation or analysis (
Our research group is active in both the Geo- and Bio-world. We will promote the eScience approach and disseminate the results through publications in both disciplines. Since 2008 we organize one or two international PhD summer schools every year. We envisage to organize a future summer school on 'OBIA of LiDAR point clouds for ecological applications'. LiDAR and OBIA also play an important role in education at the University of Amsterdam where we contribute to the eScience training of the next generation of geo-ecologists. Macroecology and RS are important and promising research directions of our permanent staff members. A follow-up with new (international) projects is thus guaranteed and as the developed infrastructure is crucial it will be maintained after the project.
Use of the national e-infrastructure
We have agreed with SurfSara (https://www.surf.nl/en/about-surf/subsidiaries/surfsara/) to use the National e-Infrastructure of the Netherlands (e-Infra). The project will require a substantial storage for all raw data and LiDAR files (max 0.5 PB). Since we have a very good experience with the central hosting of data at SurfSara, the PostgreSQL/GIS database of the proposed research will be hosted by e-Infra. In the beginning, we will collect LiDAR data and store these in a file-based archive. During the development phase, we will then use subsets of the LiDAR data from different countries. The vast majority of the LiDAR data will only be used when upscaling the analysis to Europe for relevant ecological applications (Fig.
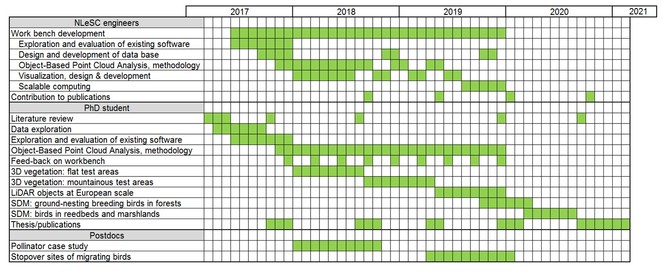
Workplan and time table
The suggested workplan and time table is illustrated in Fig.
We envisage that we need various expertise of eScience engineers. We will take advantage of the existing knowledge in the Netherlands eScience Center (NLeSC) regarding handling, storing and visualizing of LiDAR data. NLeSC knowledge and skills on machine learning and scientific visualization are essential as eScience engineers will take the lead in developing the workbench. This will be in close collaboration with the PhD student and the supervisors who will be involved in the design and will generate feed-back during the various phases in the development process. At the end of the 3rd year when OBIA is applied to large areas the PhD student will probably need some help with scalable computing. We also envisage several brainstorms with the involved eScience engineers, the PhD student, supervisors and associated researchers to design the workbench as a whole and to discuss the requirements. After three years with contributing to the methodological and technological challenges, the involvement of eScience engineers will be limited to their input to scientific publications and outreach. The PhD will then finish his/her research by using the classified objects for species distribution models. We foresee that the eScience engineers will be mainly working at NLeSC with frequent meetings (at least one per week as our institute is just across the street) with the PhD student and one of the supervisors.
Deliverables
- A SWOT (Strengths, Weaknesses, Opportunities, and Threats) analysis (provided as report), with evaluation of existing open source software that can be used for the workbench, both from the developer and user point of view.
- A file-based archive with LiDAR point cloud data
- Data base for LiDAR meta data, object models, and classified objects with their metrics, attributes and summary statistics. The data base will also include climate data, other remote sensing layers and bird data that are needed for developing species distribution models. The LiDAR objects information in the data base will be made available to the scientific community.
- Workbench focusing on Object Based Point Cloud Analysis, tested by user(s)
- PhD-thesis with 4-5 scientific publications focusing on 1) methodology of Object-Based Point Cloud Analysis (incl. case applications); 2) workbench design for Object-Based Point Cloud Analysis; 3) data publication(s) providing the classified objects from different LiDAR datasets, 4) the added value of 3D vegetation information for distribution modelling of ground nesting forest birds; and 5) identification of reedbeds from LiDAR data and its application to the distribution and abundance of birds in reedbeds and marshlands.
- PhD summer school on 'OBIA of LiDAR point clouds for ecological applications'
- Presentations at conferences and workshops and general outreach.
Acknowledgements
We thank two anonymous referees and the assessment committee of the Netherlands eScience Center for the positive evaluation of this grant proposal.
Funding program
Accelerating Scientific Discovery (ASDI) grant (ASDI.2016.014) from the Netherlands eScience Center (NLeSC)
References
- Optimizing land cover classification accuracy for change detection, a combined pixel-based and object-based approach in a mountainous area in Mexico.Applied Geography34:29‑37. https://doi.org/10.1016/j.apgeog.2011.10.010
- Susceptibility of pollinators to ongoing landscape changes depends on landscape history.Diversity and Distributions21(10):1129‑1140. https://doi.org/10.1111/ddi.12350
- Functional traits help to explain half-century long shifts in pollinator distributions.Scientific Reports6https://doi.org/10.1038/srep24451
- Tutorial: Point Cloud Library: three-dimensional object recognition and 6 DOF pose estimation.IEEE Robotics & Automation Magazine19(3):80‑91. https://doi.org/10.1109/MRA.2012.2206675
- Segmentation optimization and stratified object-based analysis for semi-automated geomorphological mapping.Remote Sensing of Environment115(12):2976‑2985. https://doi.org/10.1016/j.rse.2011.05.007
- Object based image analysis for remote sensing.ISPRS Journal of Photogrammetry and Remote Sensing65(1):2‑16. https://doi.org/10.1016/j.isprsjprs.2009.06.004
- What's wrong with pixels? Some recent developments interfacing remote sensing and GIS.GIS | Zeitschrift für Geoinformationssysteme14:12‑17.
- Progress in operational flood mapping using satellite synthetic aperture radar (SAR) and airborne light detection and ranging (LiDAR) data.Progress in Physical Geography40(2):196‑214. https://doi.org/10.1177/0309133316633570
- A multi-scale examination of stopover habitat use by birds.Ecology88(7):1789‑1802. https://doi.org/10.1890/06-1871.1
- Biodiversity loss and its impact on humanity.Nature486(7401):59‑67. URL: http://dx.doi.org/10.1038/nature11148
- Accelerated modern human–induced species losses: Entering the sixth mass extinction.Science Advances1(5). https://doi.org/10.1126/sciadv.1400253
- Habitat selection in birds.Academic Press,Orlando.
- Integrated radar and lidar analysis reveals extensive loss of remaining intact forest on Sumatra 2007–2010.Biogeosciences12(22):6637‑6653. https://doi.org/10.5194/bg-12-6637-2015
- The extent of edge effects in fragmented landscapes: Insights from satellite measurements of tree cover.Ecological Indicators69:196‑204. https://doi.org/10.1016/j.ecolind.2016.04.018
- Advances in animal ecology from 3D-LiDAR ecosystem mapping.Trends in Ecology & Evolution29(12):681‑691. https://doi.org/10.1016/j.tree.2014.10.005
- Bird migration flight altitudes studied by a network of operational weather radars.Journal of The Royal Society Interface8(54):30‑43. https://doi.org/10.1098/rsif.2010.0116
- An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems.Remote Sensing of Environment154:378‑386. https://doi.org/10.1016/j.rse.2013.07.044
- Remote sensing of snow avalanches: Recent advances, potential, and limitations.Cold Regions Science and Technology121:126‑140. https://doi.org/10.1016/j.coldregions.2015.11.001
- A science gateway for biodiversity and climate change research.8th International Workshop on Science Gateways (IWSG 2016).
- Species distribution models: ecological explanation and prediction across space and time.Annual Review of Ecology Evolution and Systematics40:677‑697. https://doi.org/10.1146/annurev.ecolsys.110308.120159
- Testing species distribution models across space and time: high latitude butterflies and recent warming.Global Ecology and Biogeography22(12):1293‑1303. https://doi.org/10.1111/geb.12078
- 3D object-oriented image analysis in 3D geophysical modelling: Analysing the central part of the East African Rift System.International Journal of Applied Earth Observation and Geoinformation35, Part A:44‑53. https://doi.org/10.1016/j.jag.2013.11.004
- Weka — A machine learning workbench for data mining.Data mining and knowledge discovery handbook.Springer,New York, NY, USA.
- Birds and habitat.Cambridge University Press,Cambridge.
- Lidar remote sensing variables predict breeding habitat of a Neotropical migrant bird.Ecology91(6):1569‑1576. https://doi.org/10.1890/09-1670.1
- Satellite remote sensing of mangrove forests: Recent advances and future opportunities.Progress in Physical Geography35(1):87‑108. https://doi.org/10.1177/0309133310385371
- Very high resolution interpolated climate surfaces for global land areas.International Journal of Climatology25(15):1965‑1978. https://doi.org/10.1002/joc.1276
- Confronting a biome crisis: global disparities of habitat loss and protection.Ecology Letters8(1):23‑29. https://doi.org/10.1111/j.1461-0248.2004.00686.x
- Improving the efficiency and accuracy of individual tree crown delineation from high-density LiDAR data.International Journal of Applied Earth Observation and Geoinformation26:145‑155. https://doi.org/10.1016/j.jag.2013.06.003
- Delineating individual trees from Lidar data: A comparison of vector- and raster-based segmentation approaches.Remote Sensing5(9). URL: http://www.mdpi.com/2072-4292/5/9/4163
- Resource diversity and landscape-level homogeneity drive native bee foraging.Proceedings of the National Academy of Sciences110(2):555‑558. https://doi.org/10.1073/pnas.1208682110
- Spatial patterns of woody plant and bird diversity: functional relationships or environmental effects?Global Ecology and Biogeography17(3):327‑339. https://doi.org/10.1111/j.1466-8238.2007.00379.x
- Bird dietary guild richness across latitudes, environments and biogeographic regions.Global Ecology and Biogeography21(3):328‑340. https://doi.org/10.1111/j.1466-8238.2011.00679.x
- Towards global interoperability for supporting biodiversity research on essential biodiversity variables (EBVs).Biodiversity16https://doi.org/10.1080/14888386.2015.1068709
- Sources and expectations for hierarchical structure in bird-habitat associations.The Condor108(1):5‑12. https://doi.org/10.1650/0010-5422(2006)108[0005:SAEFHS]2.0.CO;2
- Linking Earth Observation and taxonomic, structural and functional biodiversity: Local to ecosystem perspectives.Ecological Indicators70:317‑339. https://doi.org/10.1016/j.ecolind.2016.06.022
- Modeling forest songbird species richness using LiDAR-derived measures of forest structure.Remote Sensing of Environment115(11):2823‑2835. https://doi.org/10.1016/j.rse.2011.01.025
- LiDAR-based solar mapping for distributed solar plant design and grid integration in San Antonio, Texas.Remote Sensing8(3). https://doi.org/10.3390/rs8030247
- LiDAR remote sensing of forest structure.Progress in Physical Geography27(1):88‑106. https://doi.org/10.1191/0309133303pp360ra
- Identification of forested landslides using LiDar data, object-based image analysis, and machine learning algorithms.Remote Sensing7(8):9705‑9726. https://doi.org/10.3390/rs70809705
- On bird species diversity.Ecology42(3):594‑598. URL: <Go to ISI>://A19610906900021
- Global effects of land use on local terrestrial biodiversity.Nature520(7545):45‑50. https://doi.org/10.1038/nature14324
- Object-based analysis of airborne LiDAR data for building change detection.Remote Sensing6(11):10733‑10749. https://doi.org/10.3390/rs61110733
- Essential Biodiversity Variables.Science339(6117):277‑278. https://doi.org/10.1126/science.1229931
- Continental-scale radar monitoring of the aerial movements of animals.Movement Ecology2(1). https://doi.org/10.1186/2051-3933-2-9
- Agree on biodiversity metrics to track from space.Nature523:403‑405.
- Scale-dependent effects of landscape context on three pollinator guilds.Ecology83(5):1421‑1432. https://doi.org/10.2307/3071954
- Object-oriented mapping of landslides using Random Forests.Remote Sensing of Environment115(10):2564‑2577. https://doi.org/10.1016/j.rse.2011.05.013
- Landscape moderation of biodiversity patterns and processes - eight hypotheses.Biological Reviews87(3):661‑685. https://doi.org/10.1111/j.1469-185X.2011.00216.x
- Massive point cloud data management: design, implementation and execution of a point cloud benchmark.Computers & Graphics49:92‑125. https://doi.org/10.1016/j.cag.2015.01.007
- Segmentation of UAV-based images incorporating 3D point cloud information.Int. Arch. Photogramm. Remote Sens. Spatial Inf. Sci.XL-3/W2:261‑268. https://doi.org/10.5194/isprsarchives-XL-3-W2-261-2015
- Lidar: shedding new light on habitat characterization and modeling.Frontiers in Ecology and the Environment6(2):90‑98. https://doi.org/10.1890/070001
- The ecology of bird communities. Part 1.Cambridge University Press,Cambridge.
- High spatial resolution remotely sensed data for ecosystem characterization.Bioscience54(6):511‑521. https://doi.org/10.1641/0006-3568(2004)054[0511:hsrrsd]2.0.co;2
- Lidar sampling for large-area forest characterization: A review.Remote Sensing of Environment121:196‑209. https://doi.org/10.1016/j.rse.2012.02.001
- Remotely sensed forest structural complexity predicts multi species occurrence at the landscape scale.Forest Ecology and Management307:303‑312. https://doi.org/10.1016/j.foreco.2013.07.023
- Environmental predictors of species richness in forest landscapes: abiotic factors versus vegetation structure.Journal of Biogeographyhttps://doi.org/10.1111/jbi.12696
- Local forest structure, climate and human disturbance determine regional distribution of boreal bird species richness in Alberta, Canada.Journal of Biogeography40(6):1131‑1142. https://doi.org/10.1111/jbi.12063