|
Research Ideas and Outcomes :
Forum Paper
|
|
Corresponding author: David J Patterson (patterson.david.joseph@gmail.com)
Academic editor: Editorial Secretary
Received: 10 Oct 2022 | Accepted: 08 Nov 2022 | Published: 10 Nov 2022
© 2022 David Patterson
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Patterson DJ (2022) The scope and scale of the life sciences (‘Nature’s envelope’). Research Ideas and Outcomes 8: e96132. https://doi.org/10.3897/rio.8.e96132
|
|
Abstract
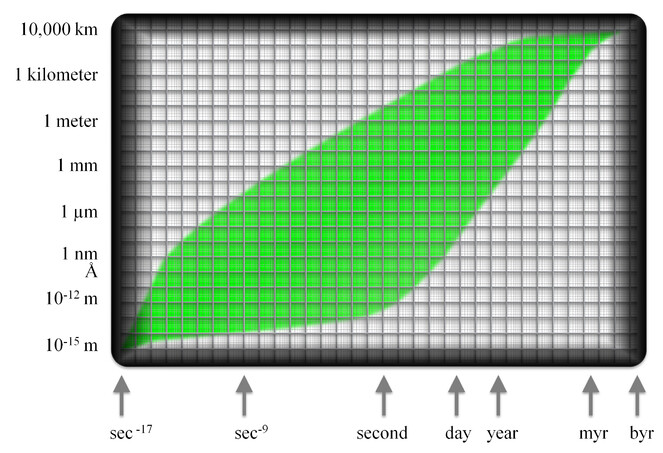
The extension of biology with a more data-centric component offers new opportunities for discovery. To enable investigations that rely on third-party data, the infrastructure that retains data and allows their re-use should, arguably, enable transactions that relate to any and all biological processes. The assembly of such a service-oriented and enabling infrastructure is challenging. Part of the challenge is to factor in the scope and scale of biological processes. From this foundation can emerge an estimate of the number of discipline-specific centres which will gather data in their given area of interest and prepare them for a path that will lead to trusted, persistent data repositories which will make fit-for-purpose data available for re-use. A simple model is presented for the scope and scale of life sciences. It can accommodate all known processes conducted by or caused by any and all organisms. It is depicted on a grid, the axes of which are (x) the durations of the processes and (y) the sizes of participants involved. Both axes are presented in log10 scales, and the grid is divided into decadal blocks with ten fold increments of time and size. Processes range in duration from 10-17 seconds to 3.5 billion years or more, and the sizes of participants range from 10-15 to 1.3 107 metres. Examples are given to illustrate the diversity of biological processes and their often inexact character. About half of the blocks within the grid do not contain known processes. The blocks that include biological processes amount to ‘Nature’s envelope’, a valuable rhetorical device onto which subdisciplines and existing initiatives may be mapped, and from which can be derived some key requirements for a comprehensive data infrastructure.
Keywords
Nature's envelope, scope of life sciences, scope of biological sciences, cyberinfrastructure, macroscope
Background
The growth of a data-rich and data-centric aspect of biology brings the prospect of new opportunities for discovery – both generally (
The potential of data-centric developments will be realized best if scientists can call on an appropriate (cyber) infrastructure that makes data freely available in a ready-to-use form. Examples of existing environments include Genbank and the other members of the International Nucleotide Sequence Database Collaboration (
The assembly of such environments for all of biology is a colossal challenge. It will be very costly and will depend on a new political commitment to fund the construction and persistence of the service infrastructure. Some argue that urgent science problems should be the driver for this new infrastructure (
The position taken here is that the research environment is ill-suited to the assembly and maintenance of a persistent service cyberinfrastructure. Most research is based on short term projects such that continued funding, and hence the continuing availability of the infrastructure, is not certain. Most current discipline-based repositories serve a particular research agenda, but lack the resources to ensure access to all data in perpetuity, provide quality control processes, or to prepare content for transfer to trusted data repositories. Without a commitment to capture all content, some legacy and at-risk information will not be made digital or not in forms that allow for easy analysis. Those data will simply be lost from inclusion in current and future scientific efforts. That is, we need to consider the needs of a comprehensive infrastructure without being constrained by what best serves trend-setters in current research.
The requirements for an ideal general-purpose (enabling) infrastructure are reasonably predictable. Using the term agent to refer to individuals, institutions, or programmes; it is expected that one or more agents will take responsibility for the discovery and aggregation of all data within each of all domains of research. The most inclusive stance should be taken as to what constitutes a domain of research. Sources should include the output of any project, individual, team, or programme; data collected by funding sources, institutions, publications, publishers, databases, computed data, and so on. Output from sources will be discovered and copied (gathered/aggregated) by agents into one or more data centres representing their defined domains of research. It is expected there will be more than 10,000 discipline-focussed data aggregators. As information may or may not have been ‘born digital’, devices will be needed to ensure that legacy data are made digital. Once acquired, data will need to be normalized, have key provenance and discipline-specific metadata added; and then be made available through reliable and trustworthy pathways for harvesting by trusted data repositories which meet CoreTrustSeal standards and which guarantee access to the data in perpetuity (
Some of the challenges that an infrastructure will face are already evident from research in biology that relies heavily on the re-use of data. A good example is the re-use of molecular data in investigations of phylogenetic relationships (e.g.
Along the pathway from source to repository, at least one agent will need to take responsibility for polishing services that will correct errors, keep metadata up to date, update software-dependent data, correct flaws in aggregation processes, and so on (
A service-oriented infrastructure must include, and be built atop, a layer of discipline-based aggregators. Absent from discussions about a general cyberinfrastructure is an assertion of the full extent of the life sciences. Such an assertion is needed to guide planning efforts with estimates of the number of data sources, the amount and character of primary data, requirements for discipline-specific data aggregation and management centers which will deliver fit-for-purpose data to persistent repositories with curatorial practices that meet the highest standards (
Nature's Envelope
The intent here is to promote the dialogue as to the scope and scale of biology that is needed to plan a data infrastructure that can serve all aspects of the biological sciences. All known life is a single array of processes which are interconnected from the sub-molecular level to the global. Each process can be represented by the size(s) of the participant(s), and its duration. Arguably, process-based metrics can be applied to any facet of biology, unlike metrics based on ‘objects’ – such as the number of species or other measures of biodiversity, the number of data objects, or the number of agents (
The graphic framework that was used for this exercise was a grid with log10 axes for the duration of processes in seconds and the size(s) of participants in metres. The choice of using a log scale is one of convenience only but is consistent with other efforts to represent information that extends over broad scales (
The result (Fig.
As biology merges with chemistry, physics, geology and other sciences, it is helpful to indicate what was included in this exercise to establish the outer bounds of the life sciences. Inclusion is limited to processes conducted by, in, or among living organisms, and the consequences of those processes. The result is not theoretical, but is a summary of processes that embrace subatomic events, molecular and biochemical events, cellular, tissue, organismic, ecological, evolutionary, and global events. Most are obviously active processes: examples being the acquisition and translocation of ions, transformational changes in motility proteins such as myosin or kinesin; the flight patterns of peregrine falcons, or the expansion of ground cover by colonial plants. Some verge on being considered passive: such as the passage of photons through chlorophyll molecules, but this is included as there is an active component that intercepts and retains energy. Also included is the expansion of the oxygen-containing atmosphere, as it is driven by photosynthetic processes. The expansion of the distribution of invasive species is included, but the movement of the virulent B.1.1.7 COVID strain aka VUI – 202012/01 recorded in Britain in October 2020, and located in the US and Australia in December of the same year is not (because of the involvement of air travel). The course of Voyager spacecraft, the ages of inert fossils, and the fossilization process are not included. Clearly, inclusion of processes as ‘life sciences’ is open to debate and may need to be reset with future versions of Nature’s Envelope.
The emphasis on processes involves an unfamiliar inexactness in information. Processes are transient by nature, are influenced by other processes, internal and external environments, recent histories, age, the number and diversity of directly or indirectly connected participants, whether information is obtained in vivo, in vitro, by inference or calculation, and so on. As an example of the imprecision involved, the time it takes for mRNA to move from a nucleus to the outer margins of a cell depends on the number of nuclear pores, the size of the cell, whether cyclosis is expressed and how, on temperature, whether the mRNA is remodelled into a ribonucleo-protein or not, involvement of molecular motors, the alleles available within the observed population, the species, and the type of cell. Consequently, the speed of movement varies by at least two orders of magnitude (
The extremes of the envelope that includes all life processes was set by identifying the processes with shortest and longest durations, and those with the smallest and largest participants. The briefest process is held to be the interception of a photon by a photopigment molecule during which energy is transferred from the photon to the photopigment. A photon of light travels at 300,000 km (3.108 metres) per second. A chlorophyll molecule measures about 2-3 nm or 3.10-9 metres. A simple calculation establishes that the amount of time that a chlorophyll molecule is exposed to and must take advantage of the energy of a photon is 10-17 seconds. As for the size of the participating photon, the treatment of photons as objects with size is questionable, but there is a consensus that a size of 1.10-15 m is appropriate (
To populate the envelope and establish its shape, sample biological processes were mapped into decadal blocks within the grid. As an example, the process of a (dead) whale exploding from pressure of gasses accumulating in its intestines endures for about 1 to 10 seconds and involves an object about 10 metres long. As with all other processes, the explosive event is not isolated. It is interconnected with the metabolism and growth of individual bacteria, populations of genetically similar organisms and of taxonomically diverse communities all of which contribute to the production of the gases. The eruption is also connected to responses by members of the microbial food web and other scavengers that benefit from the resulting supply of dissolved and particulate food materials.
Three classes of further examples illustrate the process by which the envelope was populated, and reveal more of the problems that were encountered.
Life history data are included for all classes of organisms, from sub-micron viruses to honey fungi and tree clones extending over multiple kilometres. Examples with short and long life-spans were favoured. Data on the generation times of identified bacteria measured in minutes, to various species of trees known to be many thousands of years old were included. Examples were mapped onto the decadal blocks defined by the sizes of individuals of the relevant species. Times of early demise and fossilization processes were not included. Data on life-spans were extended to classes of cells. The doubling times of many protists (single cells) are known and some were included. The life-spans of human red blood cells populate two decadal blocks. Both are defined by the size 1-10 µm (red blood cells are 7-8 µm in diameter), but given that red blood cells can survive for 70-140 days, two blocks (defined by 106 and 107seconds) were selected (
A second class of examples relate to movements. Included are the increases in dimensions of organisms from nascent form to adult. An entry for growth of stromatolites is based on estimates of a few millimetres expansion per year. For some, data are entered for a species (Arctic terns migrate more than 10,000 kms in 3 months); while others and preferred are particularized. Joe, a tumbler pigeon, departed Oregon (USA) on October 29th, 2020, and arrived 17,000 kms distant in Melbourne (Australia) on December 26th, 2020. Some activities are represented by more than one entry. Murmurations of starlings are included both in decadal blocks defined by the size of individual organisms, and in blocks for the whole flock. Cyclical movements include the molecular motor kinesin that steps 10 nm or so in 100 microseconds as well as movements of organisms from bacteria to large trees in response to tidal, diurnal, lunar, seasonal, or annual cycles. Emergence events of Magicicada that are separated by many years are included. Movements in response to environmental factors, optimising location or orientation relative to directional factors (sunlight) or to gradients (such as the responses of microbial and meiobenthic communities to REDOX gradients) are included. Range extensions are included. Cane toads were introduced to North Queensland (Australia) in 1935 with the intent of controlling pests in sugar cane crops have since expanded their range by over 1000 kms. Entries for plants include the estimated 14,000 – 80,000 year period that the Pando clone of aspen trees has extended about 5 km (
The last suite is much of a miscellany. The envelope includes transactions, such as steps in metabolic pathways, the exchange of neurotransmitters between cells, communications internally in multicellular organisms involving hormones, or externally involving pheromones. Microbial biogeochemical activities which are associated with transformations and precipitations of organic and inorganic deposits, including fool’s gold (
Concluding comments
‘Nature’s envelope’ (v. 1) is not intended as an analytical tool, but as a rhetorical device. Such devices have had a significant impact on the development of our discipline. Examples include the depiction of evolutionary relationships, the concept of evolution, so-called ‘laws’ like Gause’s Law of competitive exclusion or Bergmann’s Rule that within a clade those species that live in colder climes are larger, and various models from molecular to ecological that seek to represent reality. Although such devices may lack numeracy and exactness, they can be treated as testable hypotheses, and can grow into or spawn more exact assertions.
As a rhetorical device, Nature’s Envelope aims to provide context for a variety of conversations. Initially, it was motivated by the challenges of building a unifying informatics framework that might aid the study of any aspect of biology. It is not intended to be part of the data infrastructure. Indeed, its reliance on information about processes processes may make it incompatible with the object-based catalogues which lack information on time-lines but which are the most usual form of data repositories. None-the-less, ‘Nature’s Envelope’ can help to determine the number of discipline-based data aggregation centres that will be needed to discover, standardize and move data from primary producers into an environment where they may be freely used in computational analyses. At this time, there is not the political will nor resources to craft, build, staff and maintain a service-oriented array of data services. For the time being at least, most developments that will form part of the infrastructure will be driven by particular research technologies and agendas (
The Envelope can be made more informative by the addition of layers. Fig.
Other layers may be developed to show which areas of biology benefit from particular technologies – such as how the individual experience window can be expanded by access to microscopes. Layers may inform us about the relevance of technologies or reveal which processes are measurable and which processes must be inferred or computed. Layering can show one or more domains where communities with particular taxonomic or other skills can add value. Layering exercises that identify subdisciplines and the targets of special interest groups, will help to clarify opportunities and requirements for data interoperability. In turn, this helps to set requirements for data and metadata standards.
While the current iteration of Nature’s Envelope’ is data-based, it is inexact and incomplete. It is a preliminary assertion that, if helpful to discussions, would be improved by being fleshed out by community involvement. It would be helpful to expand and enrich this framework. More examples will help affirm the shape of the envelope. In some cases, it will be possible to import data from environments that deal with processes, such as migrations (Megamove, Movebank or the Bird Migration Explorer), cyclic processes such as seasonal emergences; life cycles, or growth. In some cases this information can be computed from object-related environments that include time-stamps as metadata. There are other definitions of 'life' which might admit more or fewer processes. Should, for example, Natur's Envelope include technology-assisted activities or exobiological assertions. Finally, there are benefits if we identify sources of arbitrariness and reduce that feature. Progress would be best done using an open collaborative community (a template is available as Suppl. material
Acknowledgements
I thank Carl Seaquist, James Patterson, Julian Partridge, and Rebecca Lynn for their comments.
References
- Multiplexed gene control reveals rapid mRNA turnover.Science Advances3:1700006. https://doi.org/10.1126/sciadv.1700006
- A specialist’s audit of aggregated occurrence records: An ‘aggregator’s’ perspective.ZooKeys305:67‑76. https://doi.org/10.3897/zookeys.305.5438
- Preserving accuracy in Genbank.Science319:1616. https://doi.org/10.1126/science.319.5870.1616a
- Nucleotide sequence database policies.Science298(5597). https://doi.org/10.1126/science.298.5597.1333b
- The biology of time across different scales.Nat. Chem. Biol3:594‑597. https://doi.org/10.1038/nchembio1007-594
- Principles and methods of data cleaning: Primary species and species-occurrence data, version 1.0. Report for the Global Biodiversity Information Facility, Copenhagen.GBI. URL: http://www.gbif.org/document/80528
- Repositories, trust, and the CoreTrustSeal.Technical Services Quarterly36(1):61‑72,. https://doi.org/10.1080/07317131.2018.1532055
- Le Macroscope: vers une vision globale.Editions de Seuil,Paris. [InFr].
- "Pando" lives: molecular genetic evidence of a giant aspen clone in central Utah.Western North American Naturalist68:493‑497. https://doi.org/10.3398/1527-0904-68.4.493
- CoreTrustSeal.VOEB-Mitteilungen71:162‑170. https://doi.org/10.31263/voebm.v71i1.1981
- Evidence for early life in Earth’s oldest hydrothermal vent precipitate.Nature543(7643):60‑64. https://doi.org/10.1038/nature21377
- Improving opportunities for new value of Open Data: Assessing and certifying research data repositories.Data Science Journal20(1):1. https://doi.org/10.5334/dsj-2021-001
- The NCBI Taxonomy database.Nucleic Acids Research40:D136-D143https://doi.org/10.1093/nar/gkr1178
- Measurement of red cell lifespan and aging.Transfusion medicine and Chemotherapy39:302‑307. https://doi.org/10.1159/000342232
- To increase trust, change the social design behind aggregated biodiversity data.Databasebax100https://doi.org/10.1093/database/bax100
- Principles for creating a single authoritative list of the world’s species.PLoS Biology18(7). https://doi.org/10.1371/journal.pbio.3000736
- BioGeomancer: Automated georeferencing to map the world's biodiversity data.PLoS Biol4:381. https://doi.org/10.1371/journal.pbio.0040381
- An overview of computational life science databases & exchange formats of relevance to chemical biology research.Comb. Chem. High Throughput Screening16:189‑98. https://doi.org/10.2174/1386207311316030004.
- A decadal view of biodiversity informatics: challenges and priorities.BMC Ecol13(16). https://doi.org/10.1186/1472-6785-13-16.
- Data integration enables global biodiversity synthesis.Proc Nationl Acad. Sci118:2018093118. https://doi.org/10.1073/pnas.2018093118
- Synthesis of phylogeny and taxonomy into a comprehensive tree of life.PNAS112:12764‑12769. https://doi.org/10.1073/pnas.1423041112
- Connecting data and expertise: a new alliance for biodiversity knowledge.Biodiversity Data Journal7:e33679. https://doi.org/10.3897/BDJ.7.e33679
- The new bioinformatics: Integrating ecological data from the gene to the biosphere.Ann. Rev. Ecol. Evol. Syst.37:519‑544. https://doi.org/10.1146/ANNUREV.ECOLSYS.37.091305.110031
- The International Nucleotide Sequence Database Collaboration.Nucleic acids research40(Database issu):33‑37. https://doi.org/10.1093/nar/gkr1006
- GenBank is a reliable resource for 21st century biodiversity research.PLOS116:22651‑22656,. https://doi.org/10.1073/pnas.1911714116
- The selection, testing and application of terrestrial insects as bioindicators.Biol. Rev. Camb. Phil. Soc73:181‑201. https://doi.org/10.1017/S000632319700515X
- A specialist’s audit of aggregated occurrence records.ZooKeys293:1‑18. https://doi.org/10.3897/zookeys.293.5111
- On the integration of a large number of life sciences web databases. In:Data Integration in the Life Sciences.Springer[ISBN3-540-21300-7].
- Powers of ten (revised).Scientific American Library[ISBN10: 0716760088]
- "gnparser": a powerful parser for scientific names based on Parsing Expression Grammar.BMC Bioinformatics18:279. https://doi.org/10.1186/s12859-017-1663-3
- A New Biology for the 21st Century.National Academies Press,Washington. URL: http://www.ncbi.nlm.nih.gov/books/NBK32509/pdf/TOC.pdf
- Cyberinfrastructure Vision for 21st Century Discovery.NSFURL: https://www.nsf.gov/pubs/2007/nsf0728/nsf0728.pdf
- Transforming science through cyberinfrastructure: NSF’s blueprint for a national cyberinfrastructure ecosystem for science and engineering in the 21st century.NSFURL: https://www.nsf.gov/cise/oac/vision/blueprint-2019
- When and where did crows learn to use automobiles as nutcrackers?Tohoku Psychological Folia60:93‑97.
- Biological databases- integration of life science data.J. Comput. Sci. Systems Biol4:87‑92. https://doi.org/10.4172/jcsb.1000081
- Biological Informatics. Final Report of the OECD Megascience Forum Working Group on Biological Informatics.OECD,Paris.
- Graduate curriculum for biological information specialists: a key to integration of scale in Biology.International Journal of Digital Curation2:31‑40. https://doi.org/10.2218/ijdc.v2i2.27
- Evolutionary informatics: unifying knowledge about the diversity of life.Trends in Ecology and Evolution27:94‑103. https://doi.org/10.1016/j.tree.2011.11.001
- Names are key to the big new biology.TREEhttps://doi.org/10.1016/j.tree.2010.09.004
- Challenges with using names to link digital biodiversity information.Biodiversity Data Journal4(e8080). https://doi.org/10.3897/BDJ.4.e8080
- BOLD and GenBank revisited - Do identification errors arise in the lab or in the sequence libraries?PLoS ONE15(4):0231814. https://doi.org/10.1371/journal.pone.0231814
- Laser spectroscopy of muonic Deuterium.Science353:669‑673. https://doi.org/10.1126/science.aaf2468
- The geomicrobiology of gold.ISME Journal1:567‑58. https://doi.org/10.1038/ismej.2007.75
- Imaging mRNA movement from transcription sites to translation sites.Semin Cell Dev Biol18:202‑208. https://doi.org/10.1016/j.semcdb.2007.02.002.
- Decentralized but globally coordinated biodiversity data.Frontiers in Big Data3:519133. https://doi.org/10.3389/fdata.2020.519133
- . Microsoft Research[ISBN ]
- Data issues in the life sciences.ZooKeys150:15‑51. https://doi.org/10.3897/zookeys.150.1766
- Pyrite formation from FeS and H2S is mediated through microbial redox activity.Proc. Natl Acad. Sci116:6897‑6902. https://doi.org/10.1073/pnas.1814412116
- Time in powers of ten. Natural phenomena and their timescales.World Scientific,Singapore. https://doi.org/10.1142/8786
- The Census of Marine Life, the Ocean Biogeographic Information System, and where do we go from here.In:Progress in Marine Conservation in Europe, 2012.BFN Scripten,Bonn. [ISBNISBN 9783896240743 3896240749].
- Filtered-Push: a map-reduce platform for collaborative taxonomic data management. In:2009 WRI World Congress on Computer Science and Information Engineering 3 731-735.IEEE Computer Society,Washington DC. [ISBN978-0-7695-3507-4]. https://doi.org/10.1109/CSIE.2009.948
- The FAIR Guiding Principles for scientific data management and stewardship.Scientific Data3:160018. https://doi.org/10.1038/sdata.2016.18
Supplementary material
A powerpoint file with an image of Nature's Envelope as submitted to RIO, with an additional editable layer as a window