|
Research Ideas and Outcomes : Project Report
|
|
Corresponding author: Hannu Saarenmaa (hannu.saarenmaa@uef.fi)
Received: 31 May 2016 | Published: 31 May 2016
© 2018 Larissa Smirnova, Patricia Mergen, Quentin Groom, Aaike De Wever, Lyubomir Penev, Pavel Stoev, Israel Pe’er, Veljo Runnel, Antonio Camacho, Timothy Vincent, Donat Agosti, Christos Arvanitidis, Francisco Bonet, Hannu Saarenmaa
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Smirnova L, Mergen P, Groom Q, De Wever A, Penev L, Stoev P, Pe’er I, Runnel V, Camacho A, Vincent T, Agosti D, Arvanitidis C, Bonet F, Saarenmaa H (2016) Data sharing tools adopted by the European Biodiversity Observation Network Project. Research Ideas and Outcomes 2: e9390. https://doi.org/10.3897/rio.2.e9390
|
|
Abstract
A fundamental constituent of a biodiversity observation network is the technological infrastructure that underpins it. The European Biodiversity Network project (EU BON) has been working with and improving upon pre-existing tools for data mobilization, sharing and description. This paper provides conceptual and practical advice for the use of these tools. We review tools for managing metadata, occurrence data, and ecological data and give detailed description of these tools, their capabilities and limitations. This is followed by recommendations on their deployment and possible future enhancements. This is done from the perspective of the needs of the biodiversity observation community with a view to the development of a unified user interface to these data – the European Biodiversity Portal (EBP). We described the steps taken to develop, adapt, deploy and test these tools. This document also gives an overview of the objectives that still need to be achieved and challenges to be addressed for the remainder of the project.
Keywords
EU BON, GEO BON, Interoperability, data standards
Introduction
The project "Building the European Biodiversity Network" (EU BON) is a European Union funded project to build the European contribution to the Group on Earth Observations Biodiversity Observation Network (GEO BON). It was funded from the end of 2012 and will continue until spring 2017. As a result of the large number of environmental changes that are occurring globally there is an increasing demand for information on these changes. Ecologists, conservationists, land managers and decision makers want clear evidence-based guidance and projections of future scenarios. This demand for information spans scales, from local planning authorities to global organizations such as the United Nations. There is a need for answers to questions about biodiversity change and the processes that are driving these changes. Yet, understanding and predicting biodiversity change is extremely difficult. Not only do these systems behave chaotically, but data are scarce and what data exists are not collected evenly, they are biased spatially, temporarily and taxonomically (
Thousands of people and organisations hold biodiversity data (cf.
It has been suggested that "biologists are joining the Big-Data Club" (
The number of biodiversity observations is growing exponentially due to the expanding number of biodiversity related initiatives and the increased use of information technology (
Mobilization and integration of data from such diverse origins is of major importance and is one of the key objectives of the EU BON project. Within the project data mobilization has required cooperation across work packages because it needs technical, social, legal and communication skills to be successful. Ideally, data will be shared in places where it is easily discovered. Such as in the data portals of the Global Biodiversity Information Facility (GBIF); Ocean Biogeographic Information System (OBIS); Long-term Ecological Research Network (LTER) and DataONE.
The number of data and metadata standards in circulation is an obstacle to potential providers of biodiversity data, but the same is also true of the diversity of software tools. Rather than adding new tools EU BON has focused on the empowerment of existing tools and standards by broadening their interoperability, connectivity and sharing capabilities.
The main challenges identified are:
- there are plenty of tools but none can, in itself, satisfy all the requirements of the wide variety of data providers.
- gaps in data coverage and quality demand more efforts in terms of data mobilization.
To fully meet the user requirements a combination of tools have been selected, which, in form of a work-flow, will mobilize data. Some of the tools are also used to further process the data, including paper publications. Within EU BON outreach campaigns and training sessions have been organized and others are planned in the future to target efforts on data mobilization where gaps have been identified.
In this document we describe the data sharing tools that have been used as part of the EU BON project. This is not a complete list of all the data sharing tools available, but a selection of some of the most important in the field (Suppl. material
Our view of the available tools for data sharing
Generic versus specialized tools for data sharing and publishing
Spreadsheets and delimited files are widely used in data management. These generic formats are often used to share tables of semi-structured data and most are well known to the community. They are easy to use and do not require the assistance of an IT specialist. From a short-term perspective these tools provide a 'quick win' for data exchange. Still, using such tools, particularly without applying clear data standards, does not promote larger scale data management, nor interoperability. The use of proprietary systems forces data into particular formats and can become an additional barrier to data sharing, reuse, persistence and accessibility. Neither do they facilitate the citation of data and the maintenance of metadata.
In order to overcome such barriers, the community has developed data sharing tools that assert common standards and structures on users. Some tools are more generic and data schema independent and thus can be used in multiple domains, while many other tools are targeted towards particular data types, applications and purposes. Tools are often developed in the context of a project or of an application, most are useful, but some need interface applications to become interoperable. Some tools include data export functions to permit sharing in standard formats. Some data publishing tools can process raw data into reports and publications for educational, decision-making and other communication purposes.
In recent years the distinction between tools for data sharing and publication are becoming less clear-cut. Technically they implement the same interoperability systems. The remaining differences are in details, such as the abilities of a tool to offer embargo and restricted access to sensitive details. In general, data sharing tools aim to facilitate progressive curation of data, while publishing tools are suited to making a stable version of data permanently discoverable and accessible.
Tools can also be categorized into those that are distributed and those that are centralized. Distributed tools are those managed by the data custodians themselves, while the centralized ones are shared repositories where the data custodians deposit their data and the management is central. Yet other tools allow a mixed approach enabling both distributed and centralized data management. They are qualified as semi-centralized with for example regional or thematic hubs.
Some tools are specific for biodiversity data types and data standards, whereas others, such as geographic information systems (GIS) are not meant specifically for biodiversity data, but are widely used in this context. Yet other tools can also handle unstructured or semi-structured data. Specialized tools make standardization easier for the ultimate user, whereas generic tools are usually easier to cope with for the data provider. A potential problem is that it is simply not possible to create data sharing tools specialized for each possible biodiversity data type (genomic, tracking, sampling, occurrence, trait, checklist etc). Data providers frequently find themselves forcing raw data into unsuitable or poorly documented data standards. Therefore, there will always be a place for both specialized and generic tools, though there may continue to be friction on the boundaries of their use, which may be resolved by using a combination of tools.
Which tools were surveyed by EU BON?
The project partners concentrated on data publishing and data sharing tools. There are also other tools, such as those for storage, data management, data capture and portals, which may also be used in data sharing workflows, but are not included in this report.
About 30 data sharing tools used in the natural history domain have been evaluated within EU BON and the results of these assessments are presented below. A summarized overview of these tools is given in supplementary files (Suppl. materials
This analysis was based on a previous summary of tools made in the framework of the projects EDIT (European Distributed Institute of Taxonomy) and SYNTHESYS (Synthesis of Systematic Resources).
Description of the tools
The EU BON project's approach was that it is better to promote and continue the development of preexisting tools, rather than creating yet new one. This approach limits the fragmentation of the infrastructural environment and leverages former investments in software and training. A detailed description of the EU BON supported tools is provided with following structure; their specifications; the adaptations that have been made and results of the testing. These are followed by recommendations on the implementation of the tool in a working environment.
These descriptions are intended to be used to produce workflows which will form an important part of the EU BON Helpdesk aiming to support the data providers in data mobilization, through the selection of suitable standards and tools enabling visualization and interpretation of the data.
GBIF Integrated Publishing Toolkit (IPT)
Summary: To publish and share biodiversity data sets and metadata through the GBIF network. It allows publication of three types of biodiversity data: i) primary occurrence data (specimens and observations); ii) species checklists and taxonomies; iii) sample-based data from monitoring programs.
Tool description:
The GBIF IPT (Integrated Publishing Toolkit) is an open source software widely used to publish and share biodiversity datasets on the GBIF network and related networks (
Enhancements by EU BON:
The release from September 10th 2015 is the version 2.3. This version has been developed together with EU BON. It is the first prototype to allow the publication of sample-based data with several uses cases from the EU BON monitoring test sites. Sample-based data are a form of data collected from surveys by environmental, ecological, and natural resource investigations. These can be one-off studies or continuous monitoring programs. Such data are often quantitative and are collected under carefully designed sampling protocols so that the results of management can be assessed or trends of populations can be detected (
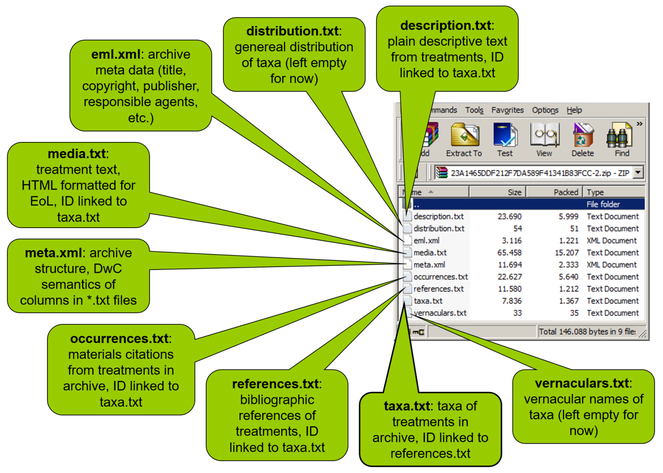
In version 2.3, a new core object, the sampling Event is introduced. The event is defined as an action that occurs at a certain location during a certain time. The IPT uses a star schema where data are connected in a one-to-many relational model. A row in the core file can be linked to many rows in one or more of the extension files. In Event data the rows of the core file contain fields common to each survey event, such as sampling protocol, sample size, sampling effort, date and coordinates. Each row in the event file is linked by a unique eventID to one or more rows in the occurence file where each row gives the details for each taxon surveyed during the event. The data can be extended with additional files to provide additional biotic and abiotic data. The schema allows the use of a “Measurement-or-facts” extension for the efficient expression of environmental information associated with the event. Or "Relevé" extension to add vegetation plot data.
The Darwin Core vocabulary already provided a rich set of terms, organized into several classes (e.g. Occurrence, Event, Location, Taxon, Identification). Many of these terms are relevant to describe sample-based data. Synthesizing several sources of input, a small set of terms relating to sample data were identified as essential, some of which were already present in the DwC vocabulary. Five new terms were ratified by TDWG (Biodiversity Information Standards) on 19 March 2015.
Specific Darwin Core terms for sample-based data (*Indicates new terms):
- eventID
- parentEventID*
- samplingProtocol
- sampleSize*
- sampleSizeUnit*
- organismQuantity*
- organismQuantityType*
Testing and implementation:
Testing of the new IPT functionalities was conducted by several EU BON partners. These tests resulted in the publication of various sample-based data from monitoring sites (
Testing was carried out using datasets from typical test site activities and included a wide range of different surveys. Doñana biological station has for instance performed tests with surveys of coastal birds. HaMaarag (Israel’s National Nature Assessment Program) tested the IPT with data from their citizen science programs (butterfly survey's and camera traps monitoring). The Rhine-Main-Observatory has published data of freshwater macrophytes and invertebrates. Marine data was covered by the Hellenic Centre for Marine Research which has published data from Amvrakikos gulf and the Sierra Nevada Global Change Observatory tested the IPT with vegetation data from forest monitoring.
Initially used as test data to prepare the training events and to test the EU BON IPT prototype, most of these data sets have been successfully published through GBIF and have enriched the biodiversity information landscape.
Future developments:
GBIF has defined next action points to enhance the latest developments, especially the introduction of the Event core (
- Monitor and report use of extension in network
- Develop visualizations to show temporal and geographic distribution of sample-based data
- Work with existing data publishers to expose additional elements from relevant datasets
- Develop filters to access data for sampling events
- Feasibility studies for further visualizations
Also tags as keywords for Essential Biodiversity Variables (EBV) classes are under consideration. There has also been discussion at Biodiversity Information Standards (TDWG) on how to develop the Darwin Core Archive (DwC-A) format further. Ontologies, such as Biological Collections Ontology (BCO), Ontology for Ecological Observational Data (OBOE), have been brought up as complementary or as an alternative (
Tool status:
An EU BON instance of the IPT is already in place at http://eubon-ipt.gbif.org together with a some test sample datasets. This IPT instance serves as the EU BON Data Repository. New versions of the IPT are available for download in both compiled and source code versions. Detailed information on how to install the tool, configure core types and extensions and publish the data can be found in the IPT user manual.
DEIMS: Drupal Ecological Information Management System
Summary: An extension of the Drupal content management system to facilitate the management and sharing of ecological data, particularly for the Long Term Ecological Research Network.
Tool description:
Drupal Ecological Information Management System (DEIMS), is a Drupal based tool to upload and share datasets providing their metadata. DEIMS is a Drupal installation profile with a set of modules and customizations for storing, editing and sharing data about biological and ecological research. It also provides web forms to describe metadata according to the Ecological Metadata Language (EML) standard. DEIMS helps the user to fill in the metadata and provide external links to the data. Each provider is responsible for maintaining the data updated and publicly accessible, depending on the sharing agreements.
Developed in partnership between the US Long Term Ecological Research Network, the University of New Mexico, the University of Puerto Rico, the University of Wisconsin, and Palantir.net, DEIMS main objective is providing an unified framework for ecological information management for LTER sites, biological stations and similar research groups.
DEIMS is not strictly a data or metadata sharing tool, as far as it is not straightforwardly deployable in each provider’s infrastructure. Rather than considering it as a tool, we can describe it as an ecological content management system, which needs a Drupal 7 instance deployed and configured properly before starting to install and configure DEIMS modules. This is indeed the main disadvantage in comparison to other metadata sharing tools: it is not easy to deploy as it requires expertise in Drupal to configure the host Drupal 7 site according to the data provider requirements.
Testing and implementation:
In the particular case of LTER Europe, they host a Drupal 7 updated infrastructure, as well as documentation, guidelines and training resources, as a main dataset repository. LTER Europe datasets are public, but the forms to create and share their metadata are only accessible to LTER sites. Some of the EU BON test sites are currently sharing datasets using LTER Europe DEIMS, which are being harvested by the broker catalog service GI-cat using the DEIMS EML harvest list.
Future developments:
As an alternative, but not accessible for the moment, DEIMS metadata could be translated into ISO-19139 metadata for Geographic Information files and shared using a GeoNetwork repository, which could also generate CSW (Catalogue Services for the Web) endpoints, consumable by GI-cat. Further tasks will be performed by LTER in reference to this alternative, in order to provide publicly accessible site for GeoNetwork, translation stylesheets and the service endpoints.
Tool status:
The platform is available at https://data.lter-europe.net/deims/. The datasets are public, but the possibility to create the forms and share the metadata are only open to LTER sites. There are however strategic plans being elaborated within Geo BON, EU BON, LifeWatch, LTER and UNESCO Man and Biosphere Sites to collaborate on these aspects.
OpenRefine, DataUp and other spreadsheet tools:
Summary: The dominant use of spreadsheets by scientists has led to the creation of tools specifically to help scientists to create standardized spreadsheets. The GBIF Spreadsheet processor is a web application to support publication of data to the GBIF. The DataUp tool has been developed by DataOne specifically to help scientists create files for archiving in a repository.
Tool description:
Scientists frequently use spreadsheets because they provide flexibility in how data can be structured. However, this flexibility also often makes the data difficult to reuse (
Proprietary formats such as those used by Microsoft Excel (e.g.,.xls,.xlsx) can be incompatible with other systems and can become obsolete when they are no longer supported (
OpenRefine is recommended for data clean-up and transformation to other formats. It has extended documentation and online supporting tutorials and videos.
DataUp has been developed by DataOne to help environmental scientists to upload files to a repository. It also includes a metadata editor. It is user friendly and allows the user to login by using Google, Facebook and Microsoft accounts. Afterwards, it gives the user the possibility of entering additional personal and professional information. Files of apparently any format can be uploaded either by dragging and dropping them into the web browser or using the the file explorer. Documentation is simple, including the name and e-mail address of the provider, the file date, title, keywords, abstract, project title and data range. An additional tab allows the user to load metadata from a file, mapping the table name, table description, field name, field description, data type, and units. DataUp is easy to use, however, the documentation is basic, and it does not allow the sampling protocol associated with data to be documented.
Recognizing that spreadsheets are a common data capture and management tools for biologists and that the Darwin Core terms lend themselves to representation in a spreadsheet, three organizations, GBIF, EOL (Encyclopedia of Life), and The Data Conservancy (DataONE project), collaborated to develop the GBIF Darwin Core Archive Spreadsheet Processor, usually just called "the Spreadsheet Processor".
The Spreadsheet Processor is a web application that supports publication of biodiversity data to the GBIF network using pre-configured Microsoft Excel spreadsheet templates. Two main data types are supported: i) occurrence data as represented in natural history collections or species observational data and ii) simple species checklists.
The tool provides a simplified publishing solution, particularly in areas where web-based publication is hampered by low-bandwidth, irregular uptime, and inconsistent access. It enables the user to convert local files to a well-known international standard using an asynchronous web-based process. The user selects the appropriate spreadsheet template, species occurrence or checklist), completes it and then emails it to the processing application which returns the submitted data as a validated Darwin Core Archive, including EML metadata, ready for publishing to the GBIF or other network
Future developments:
There are currently discussions undergoing to extend number of templates for other data types (e.g. sample-based data) and to adapt them to the new DwC terms.
Biodiversity Data Journal and ARPHA publishing platform
Summary:
Driven by the increased demand of academic community and changes to policies of governments and funding agencies, in the recent years scholarly publishing undergone serious changes, with data publishing becoming increasingly important. However, preparing data for publication is a time consuming activity that few scholars will undertake without recognition from their peers (
Tool description:
The Biodiversity Data Journal (BDJ) and associated ARPHA publishing platform are an integrated system for writing, collaborating, reviewing and publishing data and their descriptions. Unlike traditional scientific publishing workflows this system intergrates the whole process from the beginning to the end.
Not only is the BDJ a novel approach to scholarly publishing, but it also has a novel peer review system that allows much wider input from the peer community. The BDJ is an open-access journal, launched with the specific aim of accelerating mobilizing and disseminating biodiversity-related data of any kind. All structural elements of the articles, that is text, descriptions, species occurrences, data tables, etc., are treated, stored and downloaded in both human and machine-readable formats. The BDJ does not make any judgement based on the percieved impact of the data. It will publish on any taxon of any geological age from any part of the world with no lower or upper limit to manuscript size. Some examples of paper typers are:
- new taxa and nomenclatural acts;
- data papers describing biodiversity-related databases;
- local or regional checklists and inventories;
- ecological and biological observations of species and communities;
- identification keys, from conventional dichotomous to multi-access interactive online keys;
- descriptions of biodiversity-related software tools.
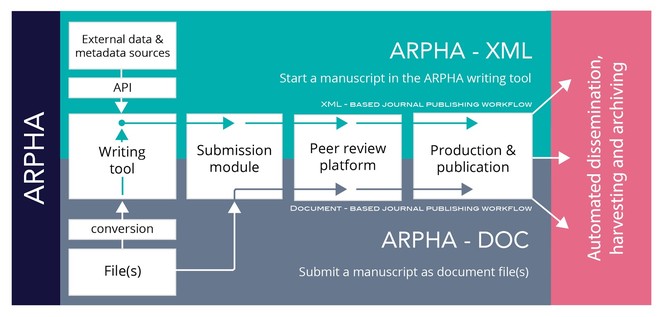
ARPHA stands for Authoring, Reviewing, Publishing, Hosting and Archiving, all in one place. It is an innovative publishing solution developed by Pensoft that supports the full life cycle of a manuscript, from authoring and reviewing to publishing and dissemination. ARPHA consists of two interconnected workflows. A journal can use either of the two or a combination of both (Fig.
The data publishing strategy of ARPHA aims at increasing the proportion of structured text and data within the article content, so as to allow for both human use and machine readability. ARPHA was successfully prototyped in 2013 by the Biodiversity Data Journal and the associated Pensoft Writing Tool. The latter, together with the document-based Pensoft Journal System (PJS), has since been upgraded, re-factored and re-branded into a generic ARPHA authoring, editorial and publishing platform. The core of this novel workflow is a collaborative online manuscript authoring module called ARPHA Writing Tool (AWT). AWT's innovative features allow for upfront markup, atomization and structuring of the free-text content already during the authoring process, import/download of structured data into/from human-readable text, automated export and dissemination of small data, on-the-fly layout of composite figures, and import of literature and data references from trusted online resources into the manuscript. ARPHA is also probably the world's first publishing system that allows submission of complex manuscripts via an API.
ARPHA provides:
- Full life cycle of a manuscript, from writing through submission, revisions and re-submission within a single online collaborative platform;
- Conversion of Darwin Core and other data files into text and vice versa, from text to data;
- Automated import of data-structured manuscripts generated in various platforms (Scratchpads, GBIF Integrated Publishing Toolkit (IPT), DataOne data base, authors’ databases);
- Automated import of occurrence data from BOLD (Barcoding of Life Databses), iDigBio (Integrated Digitized Biocollections) and GBIF platforms;
- A set of pre-defined, but flexible, Biological Codes and Darwin Core compliant, article templates;
- Easy online collaborative editing by co-authors and peers;
- A novel, community-based and public, pre-submission, pre-publication and post-publication peer-review processes.
Enhancement by EU BON:
The ARPHA Writing Tool was identified as one of the important EU BON products for data mobilization and will be incorporated into the Data publishing toolbox of the EU BON Portal.
A number of improvements of the tool were implemented as part of the project. A new plugin developed as part of EU BON to a workflow previously developed by the GBIF and Pensoft, and tested with datasets shared through GBIF and DataOne, now makes it possible to convert metadata into a manuscript for scholarly publications, with the click of a button. Pensoft has currently implemented the feature for biodiversity, ecological and environmental data. Such records are either published through GBIF or deposited at DataONE, from where the associated metadata can be converted directly into data paper manuscripts within the ARPHA Writing Tool, where the authors may edit and finalize it in collaboration with co-authors and peers and submit it to the Biodiversity Data Journal in another click.
Another new feature developed makes it possible to easily import occurrence records into a taxonomic manuscript in ARPHA. This streamlines the authoring process and significantly reduces the time needed for creation of a manuscript. Substantial amount of documented occurrence records awaiting publication are stored in repositories and data indexing platforms, such as GBIF, BOLD Systems, or iDigBio. A new upgrade of ARPHA now allows by simply specifying an identifier (ID) in the relevant box, occurrence data, stored at GBIF, BOLD systems, or iDigBio, to be directly inserted into the manuscript. It all happens in the user-friendly environment of the AWT, where the imported data can be then edited before submission to the Biodiversity Data Journal or other journals using ARPHA. Not having to retype or copy/paste species occurrence records, the authors save a lot of efforts. Moreover, they automatically import them in a structured Darwin Core format, which can be easily downloaded from the article text into structured data by anyone who needs the data for re-use after publication.
Furthermore, an automated workflow between PlutoF, which is a cloud database and computing services for Biology and related disciplines (see below) and ARPHA was established. This made possible the integration of PlutoF data into Pensoft’s ARPHA platform via an API and its subsequent publication in the Biodiversity Data Journal.
Testing and implementation:
Since its launch on 16th of September 2013 until 31 May 2016, the journal has published altogether more than 300 articles, of which 34 data papers and 10 software descriptions. The journal has got more than 1,500 users and their number increases on a daily basis.
One of the major data mobilization initiatives realized by ARPHA and BDJ is the publication of data papers on the largest European animal data base ‘Fauna Europaea’. A new series ‘Contributions on Fauna Europaea' was launched at the beginning of 2014. This novel publication model was aimed to assemble in a single collection 57 data-papers on different taxonomic groups covered by the Fauna Europaea project and a range of accompanying papers highlighting various aspects of this project (gap-analysis, design, taxonomic assessments, etc.). The first two papers were published on 17 September 2014 and until the end of 2015, 11 articles altogether have been published in BDJ (
A tutorial for the use of ARPHA called “Trips and tricks” is available on the website at: http://arpha.pensoft.net.
Tool status:
The AWT is fully operational and currently used by four Pensoft journals – Biodiversity Data Journal, Research Ideas and Outcomes, One Ecosystem and BioDiscovery. New functionalities are added continuously in line with the increased interest in publishing scientific data.
Future developments:
Enhancements of AWT and BDJ for traits data, and sample based Darwin Core compliant data sets are envisaged for the near future, as well as development and implementation of tools for visualization of genomic data. New article type templates are also scheduled, for instance IUCN compliant species conservation profile. Also, currently, the BDJ and AWT are constrained to be used mostly by the biodiversity community, so expansion to other scientific domains is in the forthcoming tasks of Pensoft IT department.
Metacat and Morpho
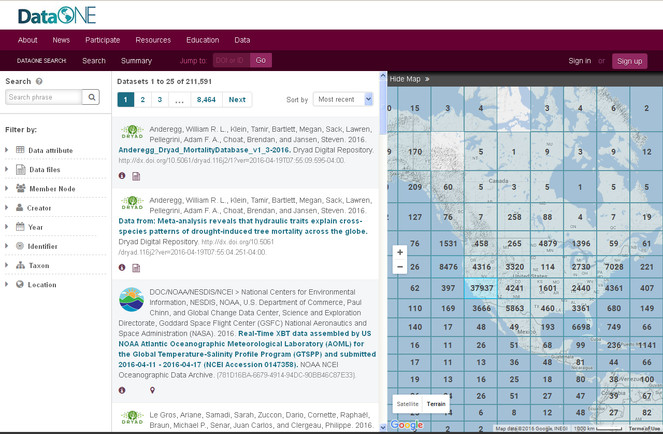
Summary: Metacat is a repository that helps scientists store metadata and data, search, understand and effectively use the data sets they manage or those created by others. A data provider using Metacat can become DataONE member node with a relatively simple configuration. Morpho is an application designed to facilitate the creation of metadata.
Tool description:
Metacat as data provider and repository
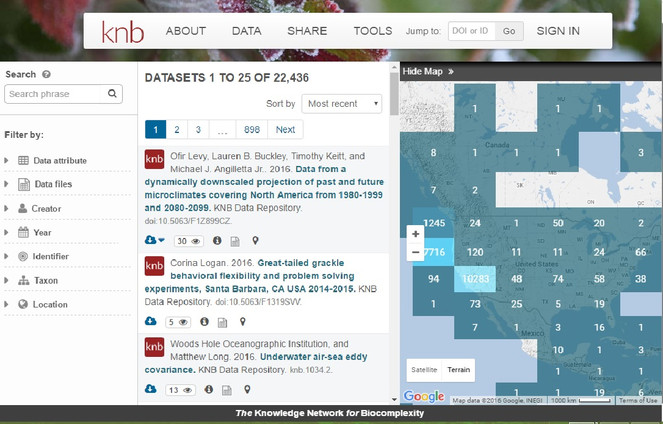
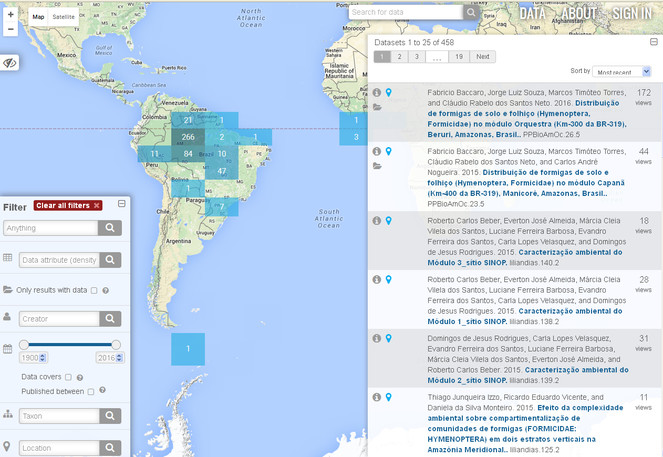
Metacat is an online database for storing ecological and biodiversity metadata and data sets. There are public Metacat repositories available for anyone to upload, search and download data and metadata or, if you have your own server available, you can install and maintain your own instance. Metacat databases can be searched by anyone using a variety of parameters (Figs
The public data repository provided by the Knowledge Network for Biocomplexity (KNB).
PPBio has installed a Metacat instance for their researchers to upload and make publicly available the results of work related to biodiversity in the Western Amazon.
Individual Metacat instances can be connected to DataOne which replicates public files. Thus the data is still available if a single instance goes offline.
Metacat is compatible with Linux, Mac OS, and Windows platforms in conjunction with a database, such as PostgreSQL (or Oracle), and a Web server. The Metacat metadata is stored in an XML format using Ecological Metadata Language (EML) or other metadata standards. Data files do not need to be compliant with any standardised data format.
Data and metadata can be entered into Metacat using an online web form (Registry application) or via Morpho, a Java program which can be downloaded and run locally. There is a user guide and a Morpho wizard that are useful for guiding providers through the process of documenting each dataset (
As data repository, Metacat allows the user to search for previously uploaded datasets using several input filters: data attributes, data files, creator, identifier, temporal coverage, taxonomic coverage and geographic coverage of the datasets. Results can be also filtered by selecting cells over a map (Figure 6).
Metacat can be configured not only as a data repository, but also as metadata provider and consumer, including support for OAI-PMH services interfaces and EML harvest lists. In particular, Metacat includes support for two OAI-PMH service interfaces: a data provider (or repository) service interface and a harvester service interface.
Testing and implementation by EU BON:
Some EU BON test sites (such as the Sierra Nevada Observatory and the Brazilian Research Program in Biodiversity) are using Metacat and Morpho for data management. In Brazil the Programa de Pesquisa em Biodiversidade (PPBio) and Programa de Pesquisas Ecológicas de Longa Duração (PELD-INPA) have set up and tested the Metacat metadata catalogue and data repository system.
Some constraines were identified:
- Flexibility that allows organizing and preserving heterogeneous datasets comes together with the drawback that it is not possible to query the data tables directly. PPBio found that it was necessary to provide auxiliary tables to allow sampling effort to be evaluated effectively in most situations.
- Effective installation can require fairly advanced knowledge of IT, and the documentation is sometimes out of date. However, the support provided by the Metacat “help-desk” is very good.
- Lack of a GitHub repository (makes user contributions to the open source slow to implement).
- There is no way to explore ecological data besides points in map.
- The process of uploading a dataset requires to manually enter the geographic coverage as latitude and logitude coordinates rather than allowing the user to select bounding boxes over a map user interface.
- Morpho does not recognize multi-domain SSL certificates.
- Use of CGI to call perl scripts in the server while logging in or registering datasets (security risk).
- DataOne cannot access Metacat servers when they are behind a proxy server.
Morpho is the default interface to upload data from desktops and is mainly used because it is necessary to check the metadata/data sent in by the researchers before it gets uploaded to Metacat, which is neccessary if the data-providers (biologists and ecologists) have no training or expe rience in data managment (
Metacat comes with a default Lightweight Directory Access Protocol (LDAP) server but it is possible to create and use your own.
GBIF is collaborating with DataONE in developing a data accessor to allow a GBIF IPT to operate independently in the DataONE network, thus bridging Metacat based datasets to the EU BON Portal. Major issues to deal with are cross mapping between metadata and preventing data replications, given that datasets are available through multiple providers.
Future developments:
The main context for use is to match the needs of EU BON as a repository for tabular data. If there are specific projects that deal with tabular data at a standardized perspective – spatial, temporal or taxonomic, it is recommended, based on PPBio experience, to build standardized data tables that will facilitate further integration. Additional development to extend the tool in order to provide a customized data-entry interface that suits the particular requirements of each project can be considered.
The Metacat tool manages to consume the same EML harvest list endpoint as DEIMS provides, but with some small differences, maybe because of the specific version of the harvest list schema (DEIMS harvest list: https://data.lter-europe.net/deims/eml_harvest_list; Metacat harvest list (from Sierra Nevada): http://linaria.obsnev.es/panel/harvestlist). Metacat instances of EU BON sites should indeed be upgraded in order to use the last EML version.
During a hackathon in Seville (26-28 January 2016), the test harvest of Granada's Metacat using its harvest list ended without success. The harvest list was compliant with EML 2.1.0 whilst GI-cat needs EML 2.1.1 compliant endpoints, due to the version of Metacat installed. The translation between both formats is feasible, e.g. using XSLT translate stylesheets, however those metadata files uploaded directly to Metacat, but not harvested, would not be published. Upgrading Metacat version in those EU BON sites using it as data repository is, therefore, advisable, in order to retrieve EML 2.1.1 compliant metadata.
As a feasible alternative to retrieve metadata from Metacat Instances, the optional Metacat OAI-PMH data provider could be installed in each test site instance. As far as GI-cat manages OAI-PMH endpoints as metadata providers, Metacat instances would be directly harvested by the GI-cat registry periodically.
Because Morpho doesn't recognize a multi-domain SSL certificate it would be more logical to replace Morpho (or having as a backup method) with Metacat's optional web-based interface for uploading data.
During a workshop in Manaus (19-25 July 2015) metadata mapping (Morpho vs. IPT) was also discussed. Within that context it may be worth considering if and what metadata fields related to systematic - monitoring schemes should be accounted for mapping Metacat/LTER datasets to EML/DwC.
Tool status:
These tools are ready to be used.
Plazi TreatmentBank and GoldenGate Imagine
Summary: A platform to store, annotate, access and distribute taxonomic treatments and the data objects within them. It works with GoldenGate and XML schemas TaxonX and TaxPub, which are tools to convert unstructured text into semantically enhanced documents with an emphasis on taxonomic data such as treatments, scientific names, material observation, traits and bibliographic references.
Tool description:
Plazi’s TreatmentBank provides access to published taxonomic treatments and their data. It also makes each treatment citable by minting persistent identifiers. Taxonomic name usages refer implicitly or explicitly to an underlying concept of the name. The taxonomic treatment includes a documentation of the traits and distribution of a related group of organisms known as a taxon (
TreatmentBank covers this niche. It offers with GoldenGate and XML schemas (TaxonX, TaxPub) open source tools to convert unstructured text into semantically enhanced documents with an emphasis on taxonomic treatments, scientific names, material observations, traits and bibliographic references (
Within TreatmentBank annotations of literature can be stored to provide links to external resources, such as specimens, related DNA samples on GenBank and literature. Annotation can be added at any level of granularity, from a material citation to detailed tagging of specimens, provision of details of the collectors and provision of morphological descriptions even to the tagging of individual traits and their states.
The use of persistent resolvable identifiers and the treatment ontology allows provision of RDF (Resource Description Framework) that supports machine harvest and logical analysis data, within and between taxa.
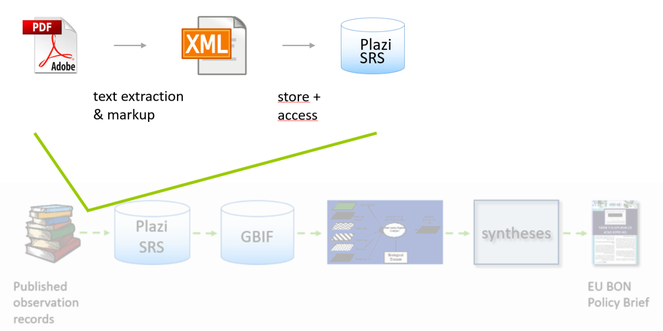
TreatmentBank provides access to data aggregators or other consuming external applications and human users, including entire treatments to the Encyclopedia of Life and observation records to GBIF using Darwin Core Archives (Fig.
Within EU BON, the GBIF pathway is the input of publication based data, specifically observation records that are linked to a treatment within an article, for EU BON’s modeling activities (Fig.
A notably value of linking TreatmentBank to GBIF and EU BON is that approximately half of the taxa are not otherwise covered within GBIF (
TreatmentBank is complemented by activities regarding legal status of treatments and other scientific facts, semantic developments, especially linking to external vocabularies and resources, and use by a number of high profile projects (GBIF, EOL, EU BON, pro-iBiosphere and some domain specific web sites). Currently 93,000 treatments from 7633 articles are available.
New technical requests can be met quickly, and Plazi has in recent years been on the forefront to build interfaces to import data into GBIF, EOL or Map of Life (i.e. DwC A). Plazi uses RefBank as a reference system for bibliographic references and is working in close collaboration with Zenodo (Biosystematics Literature Community, BLC) to build a repository for articles that are not accessible in digital form.
Future developments:
- TreatmentBank is not yet industrial strength and in its next phase it will be assess how to move from a research site to a service site.
- GoldenGate, the TreatmentBank’s central tool is powerful, but a more intuitive human-machine interface needs be developed.
- Customized versions of GoldenGate for taxonomic journals should be increased in addition to crawlers to automatically discover newly published volumes.
- Specific services need to become standalone applications, such as the parsing of bibliographic names and specimens mentioned in text.
- Chartacter trait extraction from species descriptions needs to be developed.
- TreatmentBank should become part of the LifeWatch IT infrastructure.
- In the short term, it is important to build a critical corpus of domain specific treatments to allow scientifically meaningful data mining and extraction. This may require extensive data to be gathered from treatment authors.
- Make Plazi TreatmentBank a contributor to the EU BON taxonomic backbone.
Tool status:
This tool is ready to be used. Software can be downloaded from http://plazi.org/resources/treatmentbank/.
PlutoF
Summary: An online service to create, record, manage, share, analyze and mobilize biodiversity data. Data types include ecology, taxonomy, metagenomics, nature conservation and natural history collections.
Tool description:
The PlutoF cloud provides online service to create, manage, share, analyze, and mobilize biodiversity data. Data types cover ecology, taxonomy, metagenomics, nature conservation, natural history collections, etc. Such a common platform aims to gurantee that the databases are managed within a professional, sustainable and stable architecture. It provides synergies by sharing common modules across the system. The common taxonomy module is based on standard sources such as the Fauna Europeana (
Curated datasets hosted by PlutoF cloud can be made available through public web portals. Examples include the UNITE community who curate DNA-based fungal species concepts and provide open access to their datasets through the UNITE portal. Another example is the eBiodiversity portal that includes taxonomic, ecological and genetic information on species found in Estonia. Any public dataset in the PlutoF cloud that includes information on taxa found in Estonia will be automatically displayed in this portal. This enables to have one point of access for biodiversity information on Estonia.
The implementation of mobile app tools for citizen science observations with the PlutoF API:
Community-based data generated through collaborative tools and resources are increasingly becoming a serious approach for mobilizing and generating biodiversity data for assessment and monitoring.
The PlutoF API provides a structured system that eases the implementation of citizen-science based mobile app reporting schema, thus facilitating community-based tools for data sharing. Building on the PlutoF API tools supports the primary challenge of the EU BON project to make citizen-science data qualified, available, discoverable and publicly shared.
a. Mobile app tools to support and encourage public sighting reports
Beyond the attractiveness of using state-of-the-art tools to activate the public, mobile app tools empower citizen science recording schemes and support public participation in science with a range of advantages:
- Many people across the world own mobile phones and tablets, enjoy using them, and use a range of apps (applications).
- Among the younger generation, the oppurtunity to use high-end IT tools attracts attention and interest.
- Apps are handy and easy to develop and use.
- Devices offer advanced technologies to collect and communicate valuable data in the field for enhancing data-accuracy and accurate spatial precision.
- Apps minimize effort of the user, thus, they offer an excellent tool to enhance public, voluntary participation (experts and hobbyists alike) in biodiversity reporting.
- From a policy perspective (Habitat and Birds’ Directives, EBVs), apps can facilitate rapid reporting, validation, analyses and inform policy-makers in near-real time.
- Apps broaden the range of data by allowing the collection of other types of data such as photos and sounds, which may reveal additional features such as the habitat and behavior.
b. A Citizen Science based approach for collection and qualification of biodiversity data
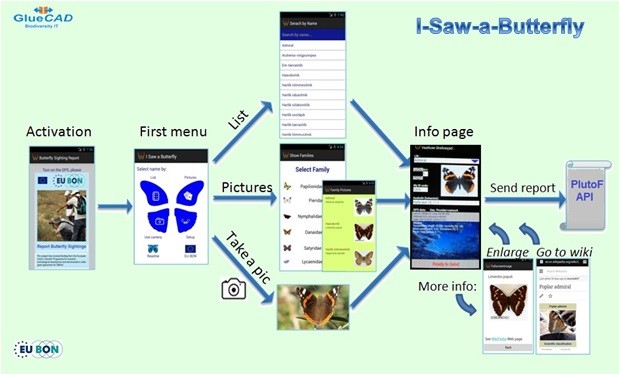
The design concept of the two mobile apps developed by GlueCAD is based on a citizen science approach that aims to take advantage of the device technology and relying less on the skill and knowledge of the user. The system supports users with data that efficiently validates and qualifies their observations. In practical terms it means relying on high-end IT devices to obtain the maximum amount of data with the minimum of typing, allowing volunteers to concentrate on observing, rather than data entry. The concept evolved getting automatic and implied data rather than relying on the skills of the user. GlueCAD has develpoped two apps one for sporadic observations and another for transects based systematic monitoring. Also, a sound recording app has been developed by the University of Tartu's Natural History Museum.
Some practical examples:
- Getting GPS information on spatial location (coordinates) as well as altitude, coordinates-accuracy and date/time for every reported species.
- Weather data can be extracted, mostly online, from nearby meteorological stations.
- Using Standard Species lists to select from.
- The speed of movement can be measured to estimate sampling-effort.
- Activation of the camera adds documents the record; may improve validation capacity and may further contribute to information about the host plants and habitat.
- Using sound recording capability of mobile devices can add multimedia content for validating observations of vocally active animals - birds, frogs, insects etc.
- A registered observer is given a user ID (which is kept in the device memory) so that there is no need to retype user details.
- Facilitation of quality control by providing information to assist validation, e.g. source of data, identified by.
- Offer observers different identification methods such as identify by list, by pictures or by voice).
c. Relying on PlutoF Taxonomic DB
Observations reported through GlueCAD's apps rely on ad-hoc querying of the API for taxon IDs, thus provides a dynamic adaptation to PlutoF, namely the standard taxonomic backbone.
It also enables future extensions to support the downloading of other taxon lists to be used for sighting reports.
d. Managing observation data with PlutoF workbench
PlutoF system allows the support of observation data moderation for any project. Observation data will then be moderated by assigned expert, before going on public display. The expert can use PlutoF's messaging interface to ask for additional information from the user to accept or decline taxon identifications for a specific observation. They can also use any additional multimedia content such as photos, videos and sound recordings to help confirm the taxon identification. Every change in the taxon identification will be recorded and can be traced within the system.
Future developments:
The University of Tartu's Natural History Museum will continue the development of the PlutoF services, partly linked to developments of national science infrastructure. New modules include water ecosystems, environmental samples, next generation sequence data, plant and forest pathology, a governmental module, and potentially a LTER module.
Tool status:
- Web-based services are available for individual users, workgroups and institutions. New infrastructure based on different technologies is under development and its beta version is available. The PlutoF Platform is developed by a team of eight software engineers.
- The mobile app for sporadic observations reporting, called “I Saw a Butterfly” is out, free, on Google Play (Fig.
7 ). Observations are recorded to PlutoF. - The second app from GlueCAD for systematic observations (“BMSapp”) is currently being tested by INPA with an Amazon’s frogs list (100) and by the Israeli group of the butterflies monitoring scheme.
Based on the range of taxonomic groups supported by the PlutoF API, it is possible to upgrade and enable the mobile app with extended lists of taxa groups for biodiversity observations recording and data sharing.
Future developments and conclusions
Challenges
Three reoccuring themes in biodiversity informatics are data openness, standardization and mobilization. All of these are pertinent to the ability to find, aggregate and use data from many sources. These are also issues which concern all of the tools mentioned in this document. Surveys have shown that scientists are still reluctant to share their data openly (
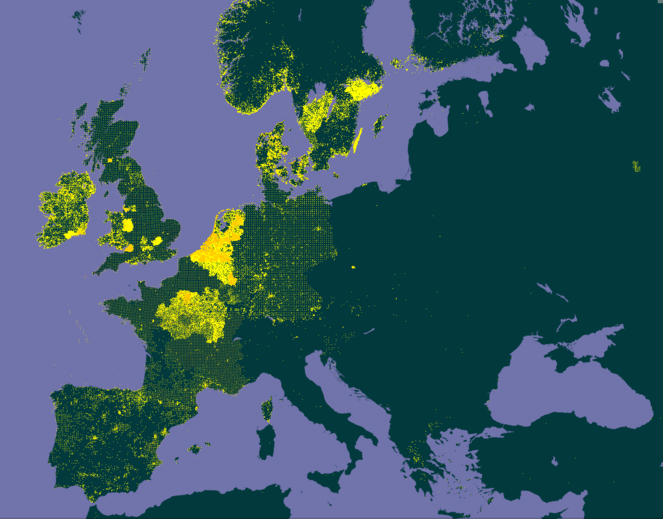
The patchiness of survey coverage in Europe illustrated by the distribution map of Plantago lanceolata taken from GBIF in 2016. This species is one of the commonest and most widespread in Europe, it should occur in almost all areas of this map, but in fact the data traces out the borders of countries and area who have published data on GBIF.
Open Data should be standard practice and should embody the principles of discoverability, accessiblity, intelligiblity and usablity (
Of paramount importance here is to collaborate on data mobilization with projects such as GEO BON, LTER and LifeWatch to provide diverse data to GBIF and to examine the advantages and problems of the new functionalities of the IPT. These may result in further recommendations and updates to the IPT. The usage of the new Darwin Core terms will also have to be followed up taking in to account of feedback from the community. This is something that will be considered by the TDWG Darwin Core working group and debated on the GBIF IPT mailing list.
Further recommendations on which data format to choose when providing data to GBIF are crucial points currently under discussion. The “Measurement Or Facts” extension was previously linked to the Occurrence Core and was used primarily to provide facts or measurements about the specimens and/or observations. The same extension linked to the Event Core allow the provision of habitat variables, parameters and descriptions. This leads to discussion on the pros and cons of the star schema approach versus using flat files and on how to interlink the different tables, as it is possible to map identical data with different core concepts. Feedback from the EU BON training events shows that potential providers have a hard time deciding which core to use, as the data are often at the borderline between Occurrences or Event Core centred datasets.
The metadata are another item of attention in the future. The sampling protocols and procedures are stored in fields in the dataset metadata. Providers are encouraged to fill them in thoroughly. By fully completing these fields, this should simplify the publishing of the dataset as a data paper. However these fields are not part of the DarwinCore terms and remain simple free text with recommendations on information to be added. They are only intended to be human-readable. In the future it would be better to have controlled vocabularies for some of these terms, such as sampling protocol.
Controlled vocabularies can be used within the DarwinCore terms to provide information on the “Gathering Event” such as including sampling methods; equipment used; information on the vessels used; the expeditions; the participating actors and the funding bodies. Data providers should be encouraged to complete both data and metadata and not to consider the human-readable metadata as a substitute of the machine-readable data which may also be needed. Thus the need for controlled vocabularies and additional terms describing the Gathering Event should be further investigated. Automated parsing or compilation of data may need to be envisaged to enhance the userfriendliness on one hand and the machine-readable requirements on the other hand.
Interesting questions were raised during training events on the IPT and on the various mailing lists on how to provide data from a monitoring scheme, where different sampling protocols were used during a same campaign in a same area. Should it be provided as several datasets each with its specific sampling protocol or can they be provided in form of one dataset listing the different sampling protocols to which the corresponding occurrences, checklists and measurement or facts should be linked to? Having a repeatable “Gathering Event” concept with associated terms as it is the case for example in the TDWG ABCD (Access to Biological Collection Data) schema could further be looked into to answer these questions.
Last but not least, questions were asked during different discussions on how to make the sample-based datasets directly discoverable when searching from the GBIF data portal, as it seems that the new terms are currently not yet indexed and thus not searchable.
In conclusion, providers and users, should be encouraged to be active in mobilizing sample-based data and to give feedback to GBIF and EU BON, so that they can be further adapted and be triggered to meet the needs and expectations of the community. There are also further needs for capacity building in data management and providing and for promoting the free and open access to data and metadata, but by repecting proper citation, re-use and compliance with national and international legislations.
Data standardization should promote analysis across much largeer areas than is currently possible by facilitating the integration of data sets. Currently, there is no central entry point for dispersed and heterogeneous biodiversity data (
Different sites are using different systems for sharing information and the challenge is trying to integrate all this information in a single metadata repository where all biodiversity information concerning EU BON appears. Future developments together with the EU BON test sites should deal with this issue, hopefully solving the limitations found in the tools that are being currently used. An analysis is foreseen of further selecting tools based on evaluation criteria for monitoring sites coupled with dedicated training in 2016.
Although the metadata language, EML, aids data discoverability, the raw data must also be accessible for automated data integration. Text and data mining tools (e.g. GoldenGate Image and Scratchpads) and further knowledge discovery would certainly help to make additional data available.
EU BON supported data sharing tools only cover some of biodiversity data types that are relevant for earth observation. Most notably, specialized tools for sharing habitat data are not yet covered. There are few such tools, as habitat data are not shared via global systems like GBIF, but some of these data can be easily exchanged using general purpose GIS and database tools. Nevertheless, the EBONE project did develop a specialized tool for habitat data, based on Microsoft Access. This tool has been evaluated, but it has been decided not to take further action, because the needs for sharing habitat data beyond what EBONE has already achieved have not yet been articulated within the EU BON project.
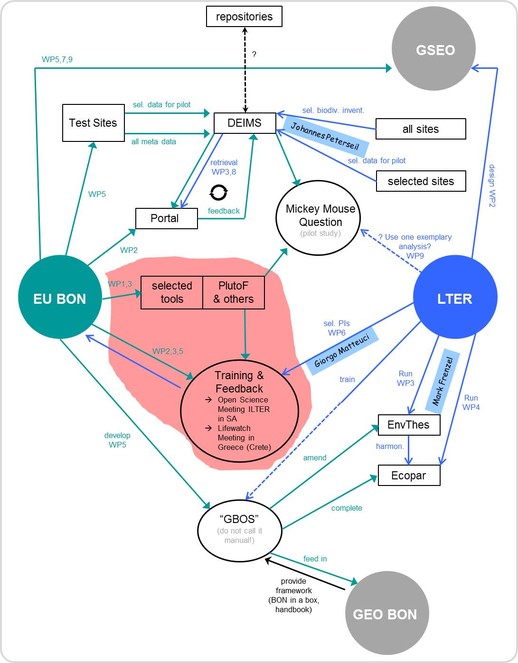
There is an agreement between EU BON and LTER to collaborate further on sharing the metadata, tools and sites among each others (Fig.
Data mobilization. EU BON is not an infrastructure project, but does have a signification infrastructure development component. As such, EU BON should devote significant resources to promote the tools and services they develop and to attract users from outside the project-funded community. The thorough gap assessment conducted by EU BON shows the most obvious temporal, spatial and taxonomic biodiversity data gaps (Fig.
To this end the EU BON commenced ongoing campaigns which should gradually lead to mobilize biodiversity data across borders, e.g. by fostering citizen science awareness and activities enforcing with guidelines towered communities that can assemble and upload their data (
For mobilizing data and promoting data sharing, EU BON has developed comprehensive training program, with a focus on data and metadata integration strategies, use of standards and data sharing tools for institutional data and IT managers, researchers, citizen scientists and monitoring programs. Several technical (informatics) workshops have been held on data standards and prototypes, e.g. of data sharing tools and the biodiversity portal. More are planned for biologists and for other life scientists from Eastern Europe who are actively involved in monitoring and managing biodiversity data.
Future development of these tools will continue to reduce obstacles to data mobilization. An enhanced workflow between data suppliers has the potential to reduce the time lag to published datasets of the huge number of data and information kept in insitutional local repositories (
These further enhancements of the tools selected for their adequacy with the objectives of EU BON will be achieved in the next steps, by involving massively the different stakeholders and outreach to additional data providers. The work done at the testing sites will now be extended to live implementation of the tools in the larger networks of the Global Biodiversity Information Facility (GBIF), Long Term Ecological Research Network (LTER), and LifeWatch, but also by encouraging smaller onganizations and individual researchers, such as those identified by the EuMon project (Biodiversity Monitoring in Europe), to use them. In this regard the helpdesk and the associated training activities will play a major role. The whole EU BON consortium is however also committed to contributing to the overall outreach efforts and is active in the implementation and enhancement of the selected data providing tools.
Funding program
The authors acknowledge the support of the EU BON project (http://www.eubon.eu), funded by the EU Framework Programme (FP7/2007-2013) under grant agreement № 308454.
Project
EU BON: Building the European Biodiversity Observation Network
Author contributions
Larissa Smirova wrote the original draft, structured the document and coordinated the authors.
Patricia Mergen helped drafting the manuscript and proof read the second draft.
Quentin Groom proof read the original report and prepared the second draft for publication.
Aaike De Wever proof reading of original report, contribution to overall manuscript and animation of initial discussions on the topic.
Pavel Stoev contributed to the Biodiversity Data Journal and ARPHA publishing platform chapter and proof read the text.
Lyubomir Penev wrote the Biodiversity Data Journal and ARPHA publishing platform chapter.
Israel Pe'er contributed to the PlutoF chapter, proof read and contributed to overall manuscript.
Velio Runnel wrote the PlutoF chapter.
Antonio García Camacho contibuted to Metacat/Morpho chapter and to overall manuscript.
Timothy Vincent contributed to the Metacat/Morpho chapter.
Donat Agosti wrote the Plazi TreatmentBank and GoldenGate Imagine chapter.
Christos Arvanitidis proof read the original report and contributed to overall manuscript.
Hannu Saarenmaa was the work package leader. He set up criteria for inclusion of the tools, determined their categorisation, wrote parts of conclusions, specified and tested the GBIF IPT enhancements, and together with Eamonn Ó Tuama worked out the new quantitative terms for Darwin Core.
References
-
Taxonomic information exchange and copyright: the Plazi approach.BMC Research Notes2(1):53. https://doi.org/10.1186/1756-0500-2-53
-
Distorted Views of Biodiversity: Spatial and Temporal Bias in Species Occurrence Data.PLoS Biol8(6):e1000385. [InEnglish]. https://doi.org/10.1371/journal.pbio.1000385
-
TaxPub: An Extension of the NLM/NCBI Journal Publishing DTD for Taxonomic Descriptions.Journal Article Tag Suite Conference (JATS-Con).Bethesda (MD): National Center for Biotechnology Information.Journal Article Tag Suite Conference (JATS-Con) Proceedings 2010,0pp. URL: http://www.ncbi.nlm.nih.gov/books/NBK47081/
-
The data paper: a mechanism to incentivize data publishing in biodiversity science.BMC Bioinformatics12:S2. https://doi.org/10.1186/1471-2105-12-s15-s2
-
Cultural Change in Data Publishing Is Essential.BioScience63(6):419‑420. https://doi.org/10.1525/bio.2013.63.6.3
-
Fauna Europaea – all European animal species on the web.Biodiversity Data Journal2:e4034. https://doi.org/10.3897/BDJ.2.e4034
-
Open exchange of scientific knowledge and European copyright: The case of biodiversity information.ZooKeys414:109‑135. https://doi.org/10.3897/zookeys.414.7717
-
Maximizing the Value of Ecological Data with Structured Metadata: An Introduction to Ecological Metadata Language (EML) and Principles for Metadata Creation.Bulletin of the Ecological Society of America86(3):158‑168. https://doi.org/10.1890/0012-9623(2005)86[158:mtvoed]2.0.co;2
-
A benchmark survey of the common plants of South Northumberland and Durham, United Kingdom.Biodiversity Data Journal3:e7318. https://doi.org/10.3897/bdj.3.e7318
-
OpenRefine (version 2.5). http://openrefine.org. Free, open-source tool for cleaning and transforming data.Journal of the Medical Library Association : JMLA101(3):233‑234. https://doi.org/10.3163/1536-5050.101.3.020
-
A decadal view of biodiversity informatics: challenges and priorities.BMC Ecology13(1):16. https://doi.org/10.1186/1472-6785-13-16
-
In Proceedings of the 14th International Conference on Scientific and Statistical Database Management, July 24-26, 2002. (2002) Key: citeulike:8241803.Fourteenth International Conference on Scientific and Statistical Database Management,Washington, DC, USA,24-26, July, 2002.Proceedings 14th International Conference on Scientific and Statistical Database Management,8pp. [ISBN0-7695-1632-7]. https://doi.org/10.1109/ssdm.2002.1029707
-
Update and Strategic Plan 2017-21.Presentation at the 13th global Nodes meeting in Antananarivo, Madagascar, 6 October 2015.GBIFURL: http://community.gbif.org/pg/file/read/50950/gnm13-session-1-update-and-strategic-plan-201721
-
Improved access to integrated biodiversity data for science, practice, and policy - the European Biodiversity Observation Network (EU BON).Nature Conservation6:49‑65. https://doi.org/10.3897/natureconservation.6.6498
-
Historical bias in biodiversity inventories affects the observed environmental niche of the species.Oikos117(6):847‑858. [InEnglish]. https://doi.org/10.1111/j.2008.0030-1299.16434.x
-
Integrating biodiversity distribution knowledge: toward a global map of life.Trends in Ecology and Evolution27(3):151‑159. [InEnglish]. https://doi.org/10.1016/j.tree.2011.09.007
-
An ontology for describing and synthesizing ecological observation data.Ecological Informatics2(3):279‑296. https://doi.org/10.1016/j.ecoinf.2007.05.004
-
Biodiversity and integrated environmental monitoring.INPA1:1‑356. URL: https://www.griffith.edu.au/__data/assets/pdf_file/0011/633962/2013-Biodiversity-and-Integrated-Env-Monitoring-BOOK.pdf
-
Mapping where ecologists work: biases in the global distribution of terrestrial ecological observations.Frontiers in Ecology and the Environment10(4):195‑201. [InEnglish]. https://doi.org/10.1890/110154
-
Biology: The big challenges of big data.Nature498(7453):255‑260. https://doi.org/10.1038/498255a
-
Integrating and visualizing primary data from prospective and legacy taxonomic literature.Biodiversity Data Journal3:e5063. https://doi.org/10.3897/bdj.3.e5063
-
Publishing sample data using the GBIF IPT (Unpublished Draft).GBIF0:1‑10. [InEnglish]. URL: http://www.gbif.org/sites/default/files/gbif_IPT-sample-data-primer_en.pdf
-
The GBIF Integrated Publishing Toolkit: Facilitating the Efficient Publishing of Biodiversity Data on the Internet.PLoS ONE9(8):e102623. https://doi.org/10.1371/journal.pone.0102623
-
Beyond dead trees: integrating the scientific process in the Biodiversity Data Journal.BDJ1:e995. [InEnglish]. https://doi.org/10.3897/bdj.1.e995
-
DataUp: A tool to help researchers describe and share tabular data.F1000Research3:6. https://doi.org/10.12688/f1000research.3-6.v1
-
Data from: Data sharing by scientists: practices and perceptions.Dryad Digital Repository1:1. https://doi.org/10.5061/DRYAD.6T94P
-
3rd EU BON Stakeholder Roundtable (Granada, Spain): Biodiversity data workflow from data mobilization to practice.Research Ideas and Outcomes2:e8622. [InEnglish]. https://doi.org/10.3897/rio.2.e8622
-
Semantics in Support of Biodiversity Knowledge Discovery: An Introduction to the Biological Collections Ontology and Related Ontologies.PLoS ONE9(3):e89606. https://doi.org/10.1371/journal.pone.0089606
-
The roles and contributions of Biodiversity Observation Networks (BONs) in better tracking progress to 2020 biodiversity targets: a European case study.Biodiversity16:137‑149.
-
Nine simple ways to make it easier to (re)use your data.Ideas in Ecology and Evolution6(2):1. https://doi.org/10.4033/iee.2013.6b.6.f
-
Darwin Core: An Evolving Community-Developed Biodiversity Data Standard.PLoS ONE7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
Supplementary materials
Download file (22.88 kb)
Definitions and concepts in the context of the main paper.
Download file (204.84 kb)
Download file (534.39 kb)