|
Research Ideas and Outcomes :
Software Description
|
|
Corresponding author: Nazareno Scaccia (nazareno.scaccia@gmail.com), Matthias Filter (matthias.filter@bfr.bund.de)
Academic editor: Editorial Secretary
Received: 15 Jun 2021 | Accepted: 04 Aug 2021 | Published: 06 Aug 2021
© 2021 Nazareno Scaccia, Taras Günther, Estibaliz Lopez de Abechuco, Matthias Filter
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Scaccia N, Günther T, Lopez de Abechuco E, Filter M (2021) The Glossaryfication Web Service: an automated glossary creation tool to support the One Health community. Research Ideas and Outcomes 7: e70183. https://doi.org/10.3897/rio.7.e70183
|
|
Abstract
Background
In many interdisciplinary research domains, the creation of a shared understanding of relevant terms is considered the foundation for efficient cross-sector communication and interpretation of data and information. This is also true for the domain of One Health (OH) where many One Health Surveillance (OHS) documents rarely contain glossaries with a list of terms for which their specific meaning in the context of the given document is defined (
New information
The Glossaryfication Web Service (GWS) is an application that automatically identifies terms in any uploaded text-based document and creates a document-specific list of matching definitions in selected online glossaries. This auto-generated document-specific glossary can easily be adjusted by the user, for example, by selecting the desired definition in case multiple definitions were found for a specific term. The document-specific glossary could then be downloaded, manually adjusted and finally included into the original document where it supports the correct interpretation of terminology used. Especially in sector-specific reports, such as from animal health or public health authorities, this can be beneficial to ensure the correct interpretation by other OH sectors in the future. The GWS was developed with the open-source desktop software KNIME Analytics Platform and runs as a web service on a KNIME Web Server infrastructure. The core data processing functionality in the GWS is based on KNIME’s Text Processing extension. KNIME's JavaScript nodes provided the basis for an interactive user interface where users can easily upload their files and select between different reference glossaries, such as the OHEJP Glossary, the CDC Glossary, the WHO Glossary or the EFSA Glossary. After retrieval of the user input settings, the GWS tags words within the provided document and maps these tagged words with matching entries in the selected glossaries. As the main output, the user receives a downloadable list of matching terms with their corresponding definitions, sectorial assignments and references, which can then be added by the user to the original document. The GWS is freely accessible via this link as well as the underlying KNIME workflow.
Keywords
One Health, OHEJP Glossary, text processing, KNIME, glossary creation
Introduction
The One Health (OH) approach aims to improve the collaboration across multiple disciplines at the national and international level in order to prevent and control emerging zoonotic diseases, as well as antibiotic resistance (
Here, we presented a new web-based solution named Glossaryfication Web Service (GWS) that enables users to automatically create document-specific glossaries by automatically searching through the OHEJP Glossary and other international glossary resources, such as CDC (Centers for Disease Control and Prevention), EFSA (European Food Safety Authority) and WHO (World Health Organisation). The GWS was designed as a user-friendly tool that adds additional value to the OHEJP Glossary and provides an easy approach to enrich future OH-related documents with more comprehensive and unambiguous glossaries. It was implemented using the open-source software KNIME (
Project description
The Glossaryfication Web Service: an automated glossary creation tool to support the One Health community
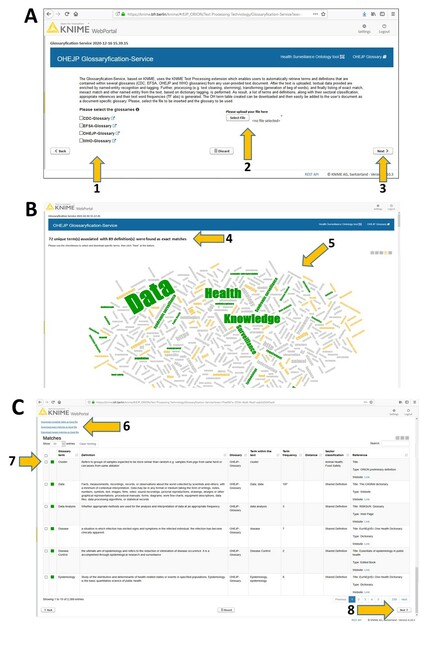
The GWS is an easy-to-use web-based tool to generate a document-specific glossary during document writing. The GWS automatically identifies nouns in a user-provided text document and yields matching definitions from the OHEJP Glossary and other supported online glossaries. Users can upload documents in different formats (e.g. PDF, Word, Excel) and choose one or more glossary(ies) to search through. Next, the GWS will automatically search within the user-provided text document for terms that are contained in the selected glossary(ies). The user will receive a downloadable list of matching glossary terms with their corresponding definitions, which can be added to the user’s document. The GWS produces a tag cloud and a table of terms with all found definitions. This table reports on exact and partly matching terms, as well as all terms for which no matching entry could be found in any of the selected online glossaries. The exact matches refer to those terms equally written in the user's provided text document and the reference glossary(ies). Inexact matches are those terms that do not exactly match the reference glossary terms, but match with small changes (e.g. plural form). The GWS provides these results in an interactive dashboard, where the end-users can select those terms and definitions that best match the intended meaning within the user-provided text document. The provided list of matching terms contains the definitions, along with their sectoral classification, references and information on the term's abundance in the provided text document. This table can easily be downloaded as an Excel file and then edited further to be finally added to the user’s document as a glossary. The GWS is accessible via regular web browsers and is operated on BfR´s KNIME Server WebPortal. On the start page of the service, a brief description is provided (Fig.
A, a screenshot of the GWS start page. B and C, output sections of the service. Specifically, B, number of exact terms and associated definitions found and, a tag cloud outcome; C, downloadable table of identified terms and definitions with term frequency, sector classification and reference. Through the GWS, end-users can upload their own file (2, A) and select which of the supported glossary(ies) should be searched through (1, A). Clicking “Next” (3, A), the GWS displays the results in an interactive table within the dashboard providing also the number of occurrences for each term in the user's document next to the identified definitions (4, B) and the tag cloud (5, B). The different colours for the terms in the tag cloud refer to different types of matches: exact matches in green, inexact matches in yellow and non-matching terms in grey. Afterwards, scrolling down the dashboard view (image C), the user has the possibility to download directly different tables (6, C) in Excel format. These options are: i) download the table with all the matches found, ii) download the exact-matches table or iii) download the inexact-matches table (6, C). Furthermore, if only a few terms with appropriate definitions are needed, the end-user can download those specific terms checking the corresponding checkboxes (7, C) and then clicking "Next" at the bottom of the dashboard page (8, C).
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No 773830.
Web location (URIs)
Technical specification
Usage licence
CC BY-NC-SA 4.0
Implementation
Implements specification
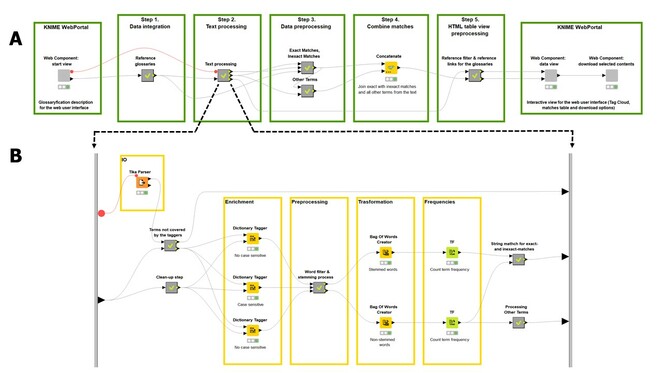
The GWS infrastructure was created with the open-source software KNIME (Konstanz Information Miner) Analytics Platform, which is a user-friendly graphical workbench for data science (
A, a screenshot of the GWS KNIME workflow. There are 5 steps within the data processing workflow, which are described in the text. The first and the last green boxes of the workflow, designed with KNIME WebPortal extension, contain so-called "Components" that provide a workflow-specific web user interface that can also be triggered by the KNIME Server. In this way, the GWS KNIME workflow becomes available as a fully functional web service in the KNIME WebPortal. B, a screenshot of the KNIME’s Text Processing extension nodes wrapped up into a metanode.
Additional information
Validation of the Glossaryfication tool and usage
The GWS was also validated using a customised text containing known terms (Suppl. material
"The following terms are found in the WHO reference glossary: Best available evidence, Bbcest available evidence, Capital investment, Capital investments, Commissioning services, Community participation, Comprehensive (maxi) HIA, Concurrent HIA, Decision making, Determinants of health, Disadvantaged / vulnerable / marginalized groups, Economic impact assessment, Employment Zone, Environmental impact assessment. Actually, some of the above terms are wrongly written."
The terms written in bold were taken from the WHO glossary and were used as "exact matches" to be retrieved by the GWS. The terms written in italic instead, represent "inexact matches" written differently on purpose for the validation exercise. All the other words from the validation text example that contain more than three letters and are not exact or inexact matches, are defined as "non-matching terms". The validation text was uploaded to the GWS via web browser and then the WHO-Glossary was selected as the reference glossary. The service found 13 exact matches out of 12. The additional exact match found is "impact assessment" that is present within the WHO reference glossary. The service finds the term "impact assessment" because it is partially identical to the WHO glossary entry "Economic impact assessment". Indeed, "impact assessment" is also recognised from the service as an inexact match. If, within a text, there is one term which is also part of a compound word that is included in the reference glossary, the compound word will appear as result of the GWS although it is not present within the text. The compound word is recognised from the service as an inexact match. The plural form of the term "Capital investment" is also retrieved as an inexact match with a Levenshtein distance of 1 due to insertion of the letter "s". The compound "Bbcest available evidence" as the inexact match is not found. This can be explained due to the insertion of more than one wrong letter in the term. The term "Bbcest available evidence" is rather divided into three different terms which appear separately as non-matching terms. If terms within the text are misspelled, those will appear as non-matching terms. In total, 27 out of 11 are obtained as non-matching terms. The high number of non-matching terms found is due to inexact matches and to the stemming process. Each word variant of an inexact match compound is reported in the list of non-matching terms. Additionally, these words, together with the other terms, are repeated twice in the list of the non-matching terms as stemmed and non-stemmed words. Examples of customised glossary tables obtained by the GWS have already been published (
Acknowledgements
This work was supported by funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No 773830: One Health European Joint Programme.
References
- KNIME - the Konstanz information miner.ACM SIGKDD Explorations Newsletter11(1):26‑31. https://doi.org/10.1145/1656274.1656280
- Characteristics of One Health surveillance systems: A systematic literature review.Preventive Veterinary Medicine181:104560. https://doi.org/10.1016/j.prevetmed.2018.10.005
- One Health Surveillance: A Matrix to Evaluate Multisectoral Collaboration.Frontiers in Veterinary Science6https://doi.org/10.3389/fvets.2019.00109
- A one health glossary to support communication and information exchange between the human health, animal health and food safety sectors.One Health13https://doi.org/10.1016/j.onehlt.2021.100263
- Towards One Health preparedness.ECDC.
- One Health Surveillance Codex: promoting the adoption of One Health solutions within and across European countries.One Health12https://doi.org/10.1016/j.onehlt.2021.100233
- The KNIME Text Processing Plugin.CiteSeerX. URL: https://www.knime.com/sites/default/files/KNIME-TextProcessing-HowTo.pdf
Supplementary material
Herein, the Glossaryfication Web Service workflow with results is provided. The workflow is already executed using a brief customised text, as described in the additional information section.