|
Research Ideas and Outcomes : Grant Proposal
|
|
Corresponding author: W. Daniel Kissling (wdkissling@gmail.com)
Received: 08 Jul 2017 | Published: 11 Jul 2017
© 2017 W. Daniel Kissling
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Kissling WD (2017) Has frugivory influenced the macroecology and diversification of a tropical keystone plant family? Research Ideas and Outcomes 3: e14944. https://doi.org/10.3897/rio.3.e14944
|
|
Abstract
Seed dispersal by fruit-eating animals is a pivotal ecosystem function in tropical forests, but the role that frugivores have played in the biogeography and macroevolution of species-rich tropical plant families remains largely unexplored. This project investigates how frugivory-relevant plant traits (e.g. fruit size, fruit color, fruit shape etc.) are distributed within the angiosperm family of palms (Arecaceae), how this relates to diversification rates, and whether and how it coincides with the global biogeographic distribution of vertebrate frugivores (birds, bats, primates, other frugivorous mammals) and their ecological traits (e.g. diet specialization, body size, flight ability, color vision etc.). Palms are particularly suitable because they are well studied, species-rich, characteristic of tropical rainforests, and dispersed by all groups of vertebrate seed dispersers. Using newly compiled data on species distributions and ecological traits in combination with phylogenies we will test (1) how fruit trait variability relates to palm phylogeny and other aspects of plant morphology (e.g. leaf size, plant height, growth form), (2) whether geographic variability in fruit traits correlates with geographic distributions of animal consumers and their traits, and (3) to what extent interaction-relevant plant traits are related to palm diversification rates. This combined macroecological and macroevolutionary approach allows novel insights into the global ecology and the evolution of a tropical keystone plant family. This is important for the conservation and sustainable management of tropical rainforests because palms are often key components of subsistence economies, ecosystem dynamics and carbon storage and therefore help to enhance nature’s goods, benefits and services to humanity.
Keywords
Frugivory, macroevolution, macroecology, biogeography, plant-animal interactions
List of participants
Research team
- Renske Onstein (Postdoc, University of Amsterdam, Amsterdam, The Netherlands)
-
W. Daniel Kissling (Associate Professor, University of Amsterdam, Amsterdam, The Netherlands)
-
William J. Baker (Head of Comparative Plant and Fungal Biology, Royal Botanic Gardens, Kew, UK)
-
Jens-Christian Svenning (Professor, Aarhus University, Denmark)
- Thomas L. P. Couvreur (Researcher, Institute of Research for Development, Marseille, France)
-
Søren Faurby (Assistant Professor, University of Gothenburg, Göteborg, Sweden)
Scientific background
A pivotal ecosystem function in tropical forests is animal-mediated seed dispersal, with about 90% of tropical plant species requiring animals to disperse their seeds (
Studies of functional and interaction diversity are almost exclusively conducted at small spatial scales, and knowledge on multispecies interactions at macroecological scales is largely lacking (
Aims, objectives and hypotheses
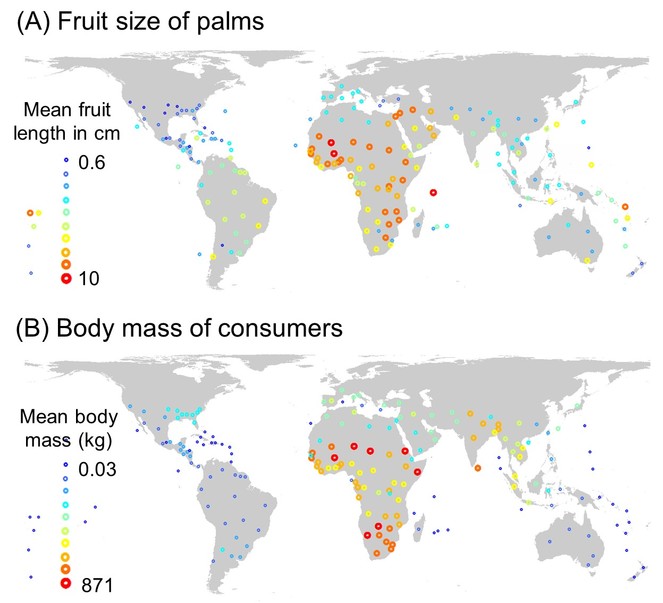
The aim of this project is to understand the macroecology and diversification of a tropical plant family in relation to frugivory-relevant plant traits and the biogeographic distribution of vertebrate consumers (Fig.
Global functional trait distributions of palms (n = 2469) and frugivorous consumers (n = 3835, incl. birds and mammals), showing large-scale co-variation between (a) palm fruit size (length in cm) and (b) consumer body size (mass in kg). Symbols are plotted for the mass centroids of geographic units ('botanical countries', TDWG level 3 units) and represent mean values across all species in a given unit. Values are calculated across a total of 29,032 species-level occurrences (5,218 for palms, 23,814 for consumers).
Using palms as a model system, the specific objectives of this project are:
- to quantify the variability of fruit traits (e.g. fruit size, colour, shape) across a tropical plant family in relation to phylogeny and other aspects of plant morphology (e.g. leaf size, plant height, growth form, etc.)
- to test how geographic variability in palm fruit traits is related to the macroecological and biogeographic distribution of avian and mammalian frugivores and their traits (e.g. body size, degree of fruit eating etc.)
- to test whether macroevolutionary diversification rates of palms are related to fruit size or other interaction-relevant traits such as plant growth form (e.g. understory vs. canopy).
These specific objectives were chosen because previous studies on plant-frugivore interactions allow to formulate a number of hypotheses about how the variability in fruit traits can be related to phylogeny, plant allometry, diversification, and geography or the biogeographic distribution of animal disperser. More specifically, the following hypotheses will be tested:
H1 (‘allometric constraints hypothesis’): Fruit traits (e.g. size, shape, colour etc.) are non-randomly distributed across phylogeny because they are phylogenetically conserved (
H2 (‘consumer biogeography hypothesis’): Geographic variability in fruit traits (size, colour, shape) is correlated with the biogeographic distribution and trait variability of animal consumers (Fig.
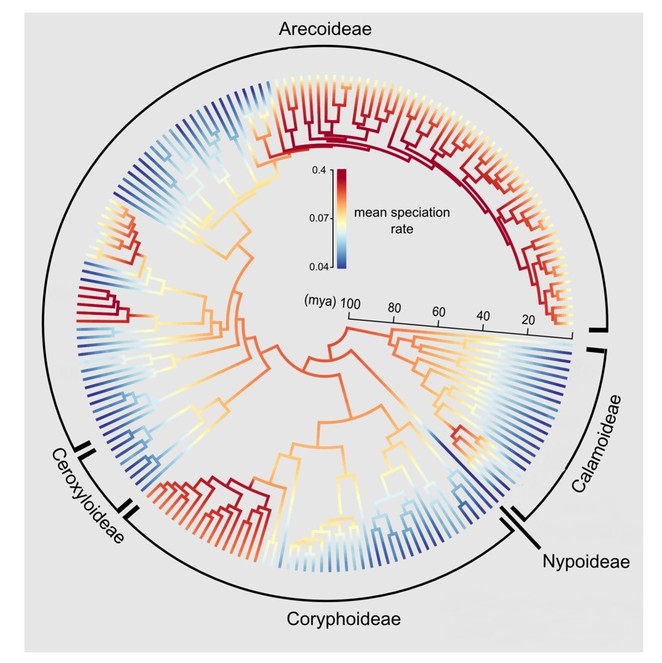
H3 (‘diversification rate hypothesis’): Fruit characteristics or other interaction-relevant traits have an influence on diversification rates (Fig.
Impact
The unprecedented and comprehensive assessment of interaction-relevant palm functional traits in relation to phylogeny, geography and animal consumers will shed new light on the evolution and biogeography of tropical rainforests. These findings will have important scientific impact by contributing to the overall assessment of what determines biodiversity (
Scientific approach
We will use a range of quantitative analytical tools for macroecological and macroevolutionary inference with large datasets, including phylogenetic comparative analyses (
Hypothesis 1: For addressing H1 (allometric constraints and variation in fruit traits), we quantify variation in fruit traits (e.g. length, width, diameter, color, shape etc.) in relation to phylogeny and allometry (leaf size, stem height, stem diameter etc.) and test for correlated evolution. We will reconstruct ancestral states of traits on the phylogenetic tree to investigate the effects of topology and branch-length variation on results. We will further test for phylogenetic signal in traits using Pagel’s λ and Blomberg’s K (
Hypothesis 2: We will implement novel co-occurrence analyses (
Hypothesis 3: We will use the newly developed Bayesian Analysis of Macroevolutionary Mixtures (BAMM;
Innovative aspects
The project has a number of unique and innovative aspects:
- Using an unprecedented global dataset on functional plant-animal traits we explore for the first time the global trait (co-)variation in palms and frugivores, a keystone tropical plant-animal interaction. To our knowledge, this dataset is exceptional as there is no other large tropical plant group for which such a complete set of resources is available. Little work has been done on quantifying functional traits at a global scale in general, and on combining trait and distribution datasets of species-rich plant and animal taxa involved in plant-frugivore interactions in particular. This will provide novel insights into the functioning of tropical ecosystems, especially into the role that mutualisms play for the origin and maintenance of tropical rainforests.
- To date, the world of macroecology and the world of macroevolution have largely operated separately. However, the great potential to combine macroecological with macroevolutionary approaches at large and temporal spatial scales has been recognized (e.g.
Jetz et al. 2012 ,Morlon 2014 ,Kissling et al. 2012b ). This offers a substantial step forward in advancing our knowledge on the drivers of biodiversity, incl. functional biogeography (Violle et al. 2014 ). We will use an integrative approach by taking advantage of cutting-edge developments in data availability, computing power and new software tools to address complex large-scale eco-evolutionary questions with informatics-based approaches. To date, nobody has applied this integrative approach to the study of plant-frugivore interactions worldwide. - Palms are a typical and important component of tropical rainforests worldwide. For instance, six out of the ten most abundant trees in the Amazon are palms (
ter Steege et al. 2013 ). Together with the availability of global datasets on species distributions, phylogeny and traits, the palm family offers a unique possibility to establish a worldwide model system for understanding the ecology and evolution of tropical rainforests and the transfer and recycling of biomass from primary producer diversity to other trophic levels (Cardinale et al. 2011 ). This is not only important from an scientific point of view, but also for the conservation and sustainable management of this highly threatened ecosystem, and the goods and services it provides to humanity.
Preliminary data
For palms, we have established a global dataset of traits covering fruit characteristics, leave size, stem height, stem diameter, climbing, and acaulescence (from herbaria, floras, monographs, botanical gardens etc.). The current dataset (February 2015, unpublished) has trait information for the majority of species (73-100% coverage) and will be finalized in this project. Distribution data of all palms have been processed before (
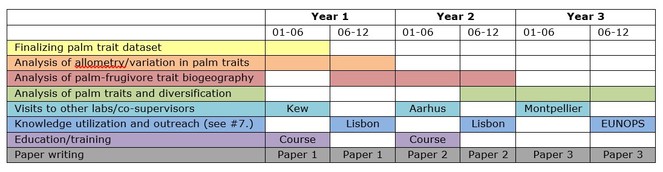
Timetable of the project
Scientific embedding of the proposed research
The applicant Dr. W. Daniel Kissling (http://www.danielkissling.de) is an internationally well-known macroecologist and biogeographer with an expertise in handling and analysis of large ecological and taxonomic datasets. In January 2014, he has been newly appointed as an Associate Professor (UHD) of Quantitative Biodiversity Science at the Institute for Biodiversity and Ecosystem Dynamics (IBED), University of Amsterdam (UvA). Dr. W. Daniel Kissling is currently establishing a new research group at IBED which aims at integrating large datasets of species distributions and ecological traits together with environmental data and phylogenies to better understand the distribution of life on Earth (the past, present, and future of biodiversity). Dr. W. Daniel Kissling currently coordinates the BSc course ‘Biodiversity & Global Change’, contributes to the UvA special research cluster 'Global Ecology', and is the scientific coordinator and PI of the newly funded Horizon 2020 project on ‘GLOBal Infrastructures for Supporting Biodiversity research (GLOBIS-B)’ (2015-2018). Dr. W. Daniel Kissling is currently a (co-)supervisor of 5 PhD students and 2 MSc students and he additionally collaborates in several large-scale biodiversity projects with postdocs, early- and late-career scientists from various countries, including Germany, Denmark, the Netherlands, Brazil, Colombia, New Zealand, the UK, and the USA. He was handling editor for the journal The Condor (2009-2010), is currently handling editor for Ecography and Journal of Biogeography, and has been appointed since 2014 to the organising committee of the Netherlands Annual Ecology Meeting (NAEM). Dr. W. Daniel Kissling has published multiple scientific papers in both multi-disciplinary scientific journals (e.g. PNAS, Proc. Roy. Soc. B, Phil. Trans. Roy. Soc B) as well as ecological high-impact journals (e.g. Biol. Rev., Ecol. Lett., Ecology, Glob Ecol. Biogeogr., J. Biogeogr., Ecography, Glob. Change Biol., etc.). He has obtained several grants (totalling ca. 2 Mio EUR) including the prestigious ‘Steno’ stipend from the Danish Council for Independent Research | Natural Sciences (similar to the Dutch VENI personal grant). Since his postdoc (2010-2011) and subsequent Assistant Professorship (2012-2013) at Aarhus University (Denmark), he has collaborated closely with three international collaborators mentioned in this proposal. Prof. dr. Jens-Christian Svenning at Aarhus University (Denmark) is an international leader in plant biogeography and has successfully established a highly productive ecoinformatics research lab with a focus on plants (including palms) and mammals. Dr. William J. Baker, Head of Comparative Plant and Fungal Biology at the Royal Botanic Gardens, Kew (UK), heads the world’s leading research group in global systematics and taxonomy of palms. Dr. Thomas L.P. Couvreur, researcher at the Institut de Recherche pour le Developpement (IRD) in Montpellier, France, is internationally known for his work on macroevolution of tropical plants, especially palms and their role for understanding the evolution of tropical rain forests. This fruitful collaboration has already resulted in several joined publications in high impact journals such as PNAS, Glob Ecology & Biogeography, Ecology, Ecology Letters, and Frontiers in Genetics. Together, the members involved in the proposed research cover a broad range of methods, experience and expertise, which will guarantee the successful implementation of the proposed research.
Knowledge utilisation
To provide the knowledge of palm traits to beneficiaries, we will collaborate with Bertrand Duval, the developer of Palmworld.org, an online platform to share detailed information about palms with non-scientific people in a playful and modern way. Palmworld is available for free on a website (http://www.palmworld.org/) as well as an iPhone/iPad App in the Apple Store (Android devices are included soon). The App has been downloaded by almost 3000 people and knowledge utilization currently amounts to 30-40 distinct people every day, totalling >13,000 accessions annually across the whole world (>180 countries). Beneficiaries include horticulturists, palm growers, field biologists, taxonomists, conservation managers, staff from botanical gardens, ethno-botanists, policy makers, people working in biotechnology, genomics and agronomy, and the general public. We will implement the species-level information about palms into Palmworld which will allow palm knowledge to be accessible in an easy and attractive format to a wide (non-scientific) audience.
In addition to providing palm trait data in Palmworld to stakeholders and the general public, the dataset will be made available to scientific research via an open-access repository. This follows the suggestions from the “datamanagementbeleid” of the NWO (letter from 9 December 2014). The proposed research further follows the standards in biodiversity informatics for accessibility and documentation of data (
Acknowledgements
Many thanks to Willem Bouten, Renske E. Onstein, William J. Baker, Jens-Christian Svenning, Thomas L. P. Couvreur and Peter H. van Tienderen for support and discussion during writing this grant proposal, and three anonymous referees for their feedback and positive evaluation.
Funding program
Open Programme (earth and life sciences) of the Netherlands Organisation for Scientific Research (NWO grant 824.15.007).
References
-
Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. I. Historical biogeography.Journal of Biogeography40(2):274‑285. https://doi.org/10.1111/j.1365-2699.2012.02795.x
-
Ecological traits influence the phylogenetic structure of bird species co-occurrences worldwide.Ecology Letters17(7):811‑820. https://doi.org/10.1111/ele.12285
-
Testing for phylogenetic signal in comparative data: behavioural traits are more labile.Evolution57(4):717‑745. https://doi.org/10.1111/j.0014-3820.2003.tb00285.x
-
The storage and production of organic matter in tropical forests and their role in the global carbon cycle.Biotropica14(3):161‑187. https://doi.org/10.2307/2388024
-
Fruit-frugivore interactions in two southern hemisphere forests: allometry, phylogeny and body size.Oikos118(12):1901‑1907. https://doi.org/10.1111/j.1600-0706.2009.17661.x
-
The functional role of producer diversity in ecosystems.Am. J. Bot.98(3):572‑592. https://doi.org/10.3732/ajb.1000364
-
Biodiversity loss and its impact on humanity.Nature486(7401):59‑67. https://doi.org/10.1038/nature11148
-
Tropical rain forest evolution: palms as a model group.BMC Biology11(1): . URL: http://www.biomedcentral.com/1741-7007/11/48
-
Origin and global diversification patterns of tropical rain forests: inferences from a complete genus-level phylogeny of palms.BMC Biology9(1):44. https://doi.org/10.1186/1741-7007-9-44
-
Global diversification of a tropical plant growth form: environmental correlates and historical contingencies in climbing palms.Frontiers in Genetics5https://doi.org/10.3389/fgene.2014.00452
-
Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families.Evolution53(3):732‑744. https://doi.org/10.2307/2640713
-
Historical contingency in the evolution of primate color vision.Journal of Human Evolution44(1):25‑45. https://doi.org/10.1016/s0047-2484(02)00167-7
-
Genera palmarum - the evolution and classification of palms.Royal Botanical Gardens,Kew.
-
Geographical ecology of the palms (Arecaceae): determinants of diversity and distributions across spatial scales.Annals of Botany108(8):1391‑1416. https://doi.org/10.1093/aob/mcr146
-
A species-level phylogeny of all extant and late Quaternary extinct mammals using a novel heuristic-hierarchical Bayesian approach.Molecular Phylogenetics and Evolution84:14‑26. https://doi.org/10.1016/j.ympev.2014.11.001
-
The ornaments of life: coevolution and conservation in the tropics.Chicago University Press,Chicago.
-
Ecological and evolutionary consequences of living in a defaunated world.Biological Conservation163:1‑6. https://doi.org/10.1016/j.biocon.2013.04.020
-
Functional extinction of birds drives rapid evolutionary changes in seed size.Science340(6136):1086‑1090. https://doi.org/10.1126/science.1233774
-
Ecology of plant speciation.Taxon59(5):1326‑1366. URL: http://www.ingentaconnect.com/content/iapt/tax/2010/00000059/00000005/art00003
-
Tempo and mode in plant breeding system evolution.Evolution66(12):3701‑3709. https://doi.org/10.1111/j.1558-5646.2012.01730.x
-
Phylogenetic inference of reciprocal effects between geographic range evolution and diversification.Systematic Biology60(4):451‑465. https://doi.org/10.1093/sysbio/syr046
-
Amazon palm biomass and allometry.Forest Ecology and Management310:994‑1004. https://doi.org/10.1016/j.foreco.2013.09.045
-
A decadal view of biodiversity informatics: challenges and priorities.BMC Ecology13(1): . URL: http://www.biomedcentral.com/1472-6785/13/16
-
Correlated evolution of fruit and leaf size in bird-dispersed plants: species-level variance in fruit traits explained a bit further?Oikos97(3):426‑432. https://doi.org/10.1034/j.1600-0706.2002.970312.x
-
Interspecific variation in fruit shape - allometry, phylogeny, and adaptation to dispersal agents.Ecology73(5):1832‑1841. https://doi.org/10.2307/1940034
-
Ecology of seed dispersal.Annual Review of Ecology and Systematics13:201‑228. https://doi.org/10.1146/annurev.es.13.110182.001221
-
Thieving rodents as substitute dispersers of megafaunal seeds.Proceedings of the National Academy of Sciences109(31):12610‑12615. https://doi.org/10.1073/pnas.1205184109
-
The global diversity of birds in space and time.Nature491(7424):444‑448. https://doi.org/10.1038/nature11631
-
Angiosperm fleshy fruits and seed dispersers - a comparative analysis of adaptation and constraints in plant-animal interactions.American Naturalist145(2):163‑191. https://doi.org/10.1086/285735
-
Spatial autocorrelation and the selection of simultaneous autoregressive models.Global Ecology and Biogeography17(1):59‑71. https://doi.org/10.1111/j.1466-8238.2007.00334.x
-
Multispecies interactions across trophic levels at macroscales: retrospective and future directions.Ecography38(4):346‑357. https://doi.org/10.1111/ecog.00819
-
The global distribution of frugivory in birds.Global Ecology and Biogeography18(2):150‑162. https://doi.org/10.1111/j.1466-8238.2008.00431.x
-
Bird dietary guild richness across latitudes, environments and biogeographic regions.Global Ecology and Biogeography21(3):328‑340. https://doi.org/10.1111/j.1466-8238.2011.00679.x
-
Cenozoic imprints on the phylogenetic structure of palm species assemblages worldwide.Proceedings of the National Academy of Sciences of the United States of America109(19):7379‑7384. https://doi.org/10.1073/pnas.1120467109
-
Quaternary and pre-Quaternary historical legacies in the global distribution of a major tropical plant lineage.Global Ecology and Biogeography21(9):909‑921. https://doi.org/10.1111/j.1466-8238.2011.00728.x
-
Establishing macroecological trait datasets: digitalization, extrapolation, and validation of diet preferences in terrestrial mammals worldwide.Ecology and Evolution4(14):2913‑2930. https://doi.org/10.1002/ece3.1136
-
Towards novel approaches to modelling biotic interactions in multispecies assemblages at large spatial extents.Journal of Biogeography39(12):2163‑2178. https://doi.org/10.1111/j.1365-2699.2011.02663.x
-
Dispersers shape fruit diversity in Ficus (Moraceae).Proceedings of the National Academy of Sciences107(33):14668‑14672. https://doi.org/10.1073/pnas.1008773107
-
Frugivore gape size and the evolution of fruit size and shape in southern hemisphere floras.Austral Ecology29(4):430‑436. https://doi.org/10.1111/j.1442-9993.2004.01382.x
-
The sizes of vertebrate-dispersed fruits - a Netropical-Paleotropical comparison.American Naturalist142(5):840‑856. https://doi.org/10.1086/285575
-
Estimating a binary character's effect on speciation and extinction.Systematic Biology56(5):701‑710. https://doi.org/10.1080/10635150701607033
-
Phylogenetic approaches for studying diversification.Ecology Letters17(4):508‑525. https://doi.org/10.1111/ele.12251
-
Plant allometry - the scaling of form and process.University of Chicago Press,Chicago and London.
-
Diversification rate shifts in the Cape Floristic Region: The right traits in the right place at the right time.Perspectives in Plant Ecology Evolution and Systematics16(6):331‑340. https://doi.org/10.1016/j.ppees.2014.08.002
-
Inferring the historical patterns of biological evolution.Nature401(6756):877‑884. https://doi.org/10.1038/44766
-
What determines species diversity.Science309(5731):90‑90. https://doi.org/10.1126/science.309.5731.90
-
Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees.Plos One9(2): . https://doi.org/10.1371/journal.pone.0089543
-
Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation.Nat Commun4https://doi.org/10.1038/ncomms2958
-
Biodiversity - Global biodiversity scenarios for the year 2100.Science287(5459):1770‑1774. URL: <Go to ISI>://000085775300030
-
Mammal predator and prey species richness are strongly linked at macroscales.Ecology94(5):1112‑1122. https://doi.org/10.1890/12-1342.1
-
Increasing awareness of avian ecological function.Trends in Ecology & Evolution21(8):464‑471. https://doi.org/10.1016/j.tree.2006.05.007
-
Gap filling in the plant kingdom—trait prediction using hierarchical probabilistic matrix factorization.Proceedings of the 29 th International Conference on Machine Learning (ICML-12)1303‑1310.
-
How colorful are fruits? Limited color diversity in fleshy fruits on local and global scales.New Phytologist198(2):617‑629. https://doi.org/10.1111/nph.12157
-
Hyperdominance in the Amazonian tree flora.Science342(6156): . https://doi.org/10.1126/science.1243092
-
Phylogenetic fields of species: cross-species patterns of phylogenetic structure and geographical coexistence.Proceedings of the Royal Society B: Biological Sciences280(1756): . https://doi.org/10.1098/rspb.2012.2570
-
The emergence and promise of functional biogeography.Proceedings of the National Academy of Sciences111(38):13690‑13696. https://doi.org/10.1073/pnas.1415442111
-
A comparison of morphological and chemical fruit traits between two sites with different frugivore assemblages.Oecologia141(1):94‑104. https://doi.org/10.1007/s00442-004-1654-8
-
The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling.Biological Reviews88(1):15‑30. https://doi.org/10.1111/j.1469-185X.2012.00235.x
-
A review of animal mediated seed dispersal of palms.Selbyana11:6‑21.