|
Research Ideas and Outcomes : Small Grant Proposal
|
|
Corresponding author: Chase C. Smith (chase.smith@mcphs.edu)
Received: 24 Jul 2016 | Published: 27 Jul 2016
© 2016 Chase C. Smith, Matthew Todd, Luc Patiny, Christopher Swain, Christopher Southan, Alice E. Williamson, Alex M. Clark.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Smith C, Todd M, Patiny L, Swain C, Southan C, Williamson A, Clark A (2016) SCINDR - The SCience INtroDuction Robot that will Connect Open Scientists. Research Ideas and Outcomes 2: e9995. doi: 10.3897/rio.2.e9995
|

|
Keywords
Open Science, Malaria, Drug Discovery, Electronic Laboratory Notebook, Reaction Informatics, Chemical Structure Alerts, Collaboration
Abstract
This project will develop a way to connect, in real time, globally disparate researchers who are doing similar science so that they can work better and faster towards the development of new medicines.
The scientific literature already fulfills the role of notifying researchers about work that has been done, and social media has recently evolved to alert researchers to what is being done. While these new communication technologies simplify the collaborative process between widely distributed researchers, there still exists a major gap in efficient real time alerting and updating. We aim to automate an alert process so that, as a researcher records what they are doing in a natural way, they are immediately alerted to others around the world in real time who are working on related science.
Our system is built on the conceptual model of the machine understanding of human-generated content, used by social media platforms to generate alerts to further relevant content. The system we propose to build will understand the molecular information being recorded in a scientist’s notebook. It will then search both its own records and others in the public domain in order to introduce scientists where there may be mutual advantage - when two laboratories are working on similar molecules, assays or approaches, for example. To achieve this, we will build on a recently developed open source electronic lab notebook (ELN) to create the required component - the automated alerting service we call the SCience INtroDuction Robot, or SCINDR.
We foresee wide application of SCINDR in chemical and biological research because it will accelerate research by connecting people. In so doing, SCINDR will provide the incentive for others to take their research into the public domain (
Project Goals for the Advancement of Open Science
The term “open science” is interpreted in many ways, but at its heart it refers to a level of transparency that permits and encourages serendipity - the chance that one’s data can be advantageously used by others, or that one’s research can be easily discovered and combined in new and unexpected ways. Collaboration lies at the centre of open science, particularly collaborations that are created by the openness of the process. The benefits of this kind of cross-fertilisation has been described the "The Medici Effect," a termed coined by Johansson proposing that "innovation comes from diverse industries, cultures, and disciplines when they all intersect, bringing ideas from one field into another [
When working openly, we pour data and ideas into the public domain, and hope that we can find and work with others to make our research better, faster or more robust. Yet we take few steps to maximize our chances of finding the right people - some may not even normally reside within the same professional circles of interaction and collaboration. For those who have worked openly, the difficult experience of finding people working on related science can be daunting, despite the productivity gains that could be realized when it does work. Can we do better? Can we automate the process? Can we help find and connect the most relevant and productive collaborators?
In the area of health research, the importance of working with the right people makes tangible differences to the lives of whole populations. Research being conducted towards new therapeutics for malaria may unearth a tool for the better understanding of dementia or a new medicine to treat the Zika virus. It would be a tragedy if such a laboratory was working in isolation and ignorance of the other. How frustrating for the patient community, who would see only wasted time and money spent on teams unable to work together by sharing their expertise and their data (both positive and negative.) Transforming this situation may involve learning a lesson from our non-science friends in advertising. In our everyday lives, we are matched with relevant content through algorithms. When we write emails, advertisements often appear that are eerily relevant to what we are reading or writing. A machine has understood the text, and is able to offer relevant and related information. Automated AI services like ‘Siri’ and ‘Google Now’ perform the same function, and music streaming services have become effective at suggesting relevant music to us, not by understanding musical theory, but by monitoring our playlists.
We propose to exploit this automated introduction service (SCINDR) to improve the efficiency of open science, in the area of health research. The underlying platform is a new open source electronic lab notebook (ELN) that we have recently built and are continually road-testing and evolving. The ELN was initially developed using open source cheminformatics tools at the Swiss Federal Institute of Technology (http://www.cheminfo.org/#). The basis of how SCINDR relates to the ELN is shown in the Supplementary Video (
The process would begin by having the health researcher first recording their data in the most natural and familiar way possible. The next step would involve the ELN storing this information in a machine understandable form in an openly accessible format. SCINDR, which we are proposing to develop, would then parse the information and search both its internal research records and those residing in publicly available web databases such as PubChem. The system would identify the connections between similar molecules, chemical reactions, biological assays or other features of importance in health research and provide real-time feedback to the scientists so that they may, if they choose, contact each other in order to begin to work together.
Our primary objective is to construct a prototype of this system as a proof of concept. This would represent an important advance; allowing real time, and automatic, connections to be established between researchers who are working on related scientific projects. Data stored in the ELN will be in the open: the efficiency benefits SCINDR will provide will create a positive incentive towards open science.
Our initial project goals are:
-
Develop and implement the required software extensions for the ELN to provide real time molecular structure alerts.
-
Deploy the upgraded ELN and pressure test the system with data originating from several existing research groups in an open environment.
-
Disseminate the ELN and newly developed code to the open community.
-
Initiate work to improve and expand the system beyond the initial roll out based on user feedback.
The initial deployment of SCINDR will be focused on identifying and alerting for molecular similarity, since this is feasible in the short term. Extensions planned will upgrade SCINDR to allow: i) chemical reaction searching; ii) protein sequence similarity for more biologically-centered queries and iii) relationships between chemical structure and natural-language descriptors, i.e. alerts that identify occurrences of molecules with particular terms (e.g., [structure X] + [the word “Zika”).
Impact
The system will be road tested initially by a community of researchers known as Open Source Malaria (OSM), a consortium funded to carry out drug discovery and development for new medicines for the treatment of malaria.
OSM is a suitable sandpit for evaluation of SCINDR. The community of researchers already exists and is continually growing. The application to malaria is significant, given the tragic cost of the disease: an estimated 214 million cases in 2015, which resulted in the deaths of 438,000 people, mostly children (
i) how to find other scientists working on related molecules, or on the syntheses of those molecules (e.g. the current “Series 4” that is OSM’s most promising set of compounds to date (
ii) those working with related structures on other diseases.
iii) those working with related biological assays/targets/hypotheses.
The impact on the OSM Project will be significant, since the identification of suitable collaborators has consumed significant amounts of project time. Our pilot studies will involve the following specific steps:
i) Validation of direct entry of chemical reaction information by researchers using an intuitive interface that provides machine-understandable data as the output (i.e. the outputs are standard file formats such as molfile, RXN, Jcamp-dx.)
ii) Comparison of the data with other pages in the ELN.
iii) Comparison of the data with open public databases, the open scientific literature and the use of Google searches (e.g. with powerful machine-understandable informatics strings such as the InChiKey.)
iv) Creation of an alert in the browser window detailing the nature and extent of the overlap and a means of contacting the person or accessing the resource.
This demonstration of SCINDR in a real project will be significant psychologically, given that the increases in research efficiency it will provide come by virtue of the research process being conducted in the open. The tangible demonstration of superior performance, rather than abstract mandates, are essential if we are to energise the transition of the science community to open methods.
This proposal describes a pilot project. When fully developed, SCINDR will be a crucial automated assistant available to work in the background of every open science project related to health research, and its combination with an intuitive ELN will be a powerful tool for the open community of health researchers, in both developed and developing countries. Community-based improvements will be sought continuously. As SCINDR develops there are a number of sustainability options for future development that would be explored, including the Red Hat software as a service model, or the pay-for-secrecy Github model.
Innovation and Originality
(The potential for automatically connecting relevant people and/or matching people with commercial content currently dominates much of software development, yet the analogous idea of automatically connecting people who are working on similar science in real time does not exist. This extraordinary fact arises in part because so few people work openly, meaning almost all the research taking place in laboratories around the world remains behind closed doors until publication (or in a minority of cases deposition to a preprint server), by which time the project may have ended and researchers have moved on or shelved a project. As open science gathers pace, and as thousands of researchers start to use open records of their research, we will need a way to discover the most relevant collaborators, and encourage them to connect. SCINDR will solve this problem.
The innovation centers around the following key features of the system:
-
Real time alerting and notification of reaction and molecular entries that overlap with similar scientific pursuits. The degree of similarity leading to an alert can be set numerically by the user to generate the desired quantity of information.
-
Ease of use. Familiar web based interface suitable for novice users with the option to modify the modular set-up for use by experts, including options to switch SCINDR on and off, and to de-prioritise those connections the user deems uninteresting.
-
Open source system with minimal IT resources required, enabling more globally dispersed researchers to take part in the discovery of medicines.
Based on the reactions and products being worked on, the page will query various databases outside the ELN to look for related content. Those databases will initially include PubChem, ChemSpider, and ChEMBL as well as being expanded to include IUPAC International Chemical Identifier Key (InChI-Key) web searches (
Technical Viability
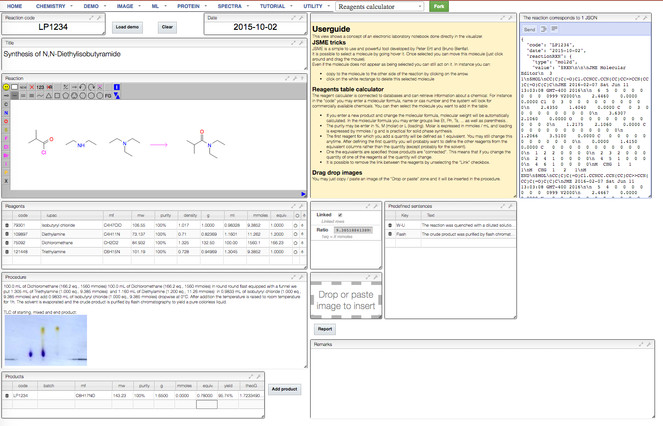
SCINDR will be built on a newly-developed ELN with which we have extensive experience, providing a strong foundation for the creation of a new tool for the community; the ELN’s creator is one of the applicants and will supervise the activities of the software coder to be employed. The ELN is a multi-user environment that can track revision history, attach and store large spectral data files and search chemical reaction information. Analytical results associated with any molecule can be retrieved and processed in real time. The ELN will be universally accessible by utilizing modern web browsers as the user interface. The database can be located remotely on a partner server or installed locally if particular labs have sufficient local IT support and resources. A prototype of the new ELN (
Example of currently functioning Electronic Laboratory Notebook (ELN) entry to which SCINDR will be added. Further enhancements such as real time NMR, LCMS and IR data visualization have been developed and will be added to extend the ELN capability.
The above mentioned ELN is the perfect platform for the addition of SCINDR since it is already acting as a repository of open drug discovery information that can be mined by the robot. SCINDR can then easily perform its similarity searches by accessing other open databases in order to make the requisite connections. Technically, SCINDR will be an open-source javascript library that will be easy to integrate in any webpage. It will call a list of predefined providers like PubChem, Chemspider and the ELN itself and will be easy to maintain. Results coming from various sources can be straightforwardly combined and ranked to yield a report in a customizable format. A video demonstrating the capability of the ELN has been included in this proposal as a supplementary video or can also be viewed on YouTube (https://www.youtube.com/watch?v=joarvBnTQ_k).
Developing SCINDR is technically viable within an initial 6-9 month window. A prototype of the ELN is already working, and chemical similarity search APIs are available for the relevant online database searches. The complete system needs to be assembled and then debugged and tested under real world conditions.
Resource Viability
The creation of SCINDR requires the resources listed in the Budget Section. The main resources required are coding time and expertise, coupled with computing and server costs, and these constitute the main budget items. The ELN code development and initial hosting is planned to occur primarily at the Ecole Polytechnique Fédérale de Lausanne, Switzerland (EPFL) in consultation with co-applicants who are chemical software experts from the Biotech / Pharmaceutical sector. The amount requested is appropriate for a full-time software developer for 6 months based at the EPFL.
Medicinal Chemistry groups from the United States (MCPHS University, School of Pharmacy, Worcester, MA, USA) and Australia (School of Chemistry, The University of Sydney, Australia) will test the robustness of the system by generating real data from the OSM Project and provide feedback for further improvements and modifications to the software team for real world deployment. OSM has contributors from multiple countries (Australia, United States, Sweden, Switzerland, United Kingdom, India and Canada), each providing their unique resources and experience in open channels.
The funding for the Research Student is to pay for their time as a project coordinator: disseminating SCINDR to the wider community, collating feedback and contacting groups with potential interest in the outcome of the project. The student will also write up one blog post per month during the opening phase of the project to summarise progress and needs.
Travel and meeting costs are requested for 2 members of the team to travel to 2 relevant scientific conferences during Phase I to present the project to relevant communities. Publication costs are requested because an output of the initial phase will be a peer-reviewed publication in an open access journal co-authored by the team and any community contributors.
Laboratory equipment and consumables costs are required for PI Smith’s laboratory in order to carry out research into new antimalarials that will be used as the test-bed for the performance of SCINDR. These costs in the Sydney laboratory will be met from existing grant money.
Budget
See
Estimated Budget for SCINDR Project.
|
Budget Category |
Cost Estimate |
|
Salary Costs for Coding Development |
$50,000.00 |
|
Associated Server / Computing Costs |
$5,000.00 |
|
Travel Costs |
$6,000.00 |
|
Publication Costs |
$1,000.00 |
|
Meeting Costs |
$3,000.00 |
|
Research Student Stipends |
$5,000.00 |
|
Laboratory Equipment Costs |
$5,000.00 |
|
Laboratory Consumables Costs |
$5,000.00 |
|
Total: |
$80,000.00 |
References
-
Medici Effect: What Elephants and Epidemics Can Teach Us about Innovation.Harvard Business Review Press,Boston,224pp. [InEnglish]. URL: http://www.fransjohansson.com/the-medici-effect-by-frans-johansson/ [ISBN9781422102824]
-
OpenSourceMalaria:Triazolopyrazine (TP) Series. http://openwetware.org/wiki/OpenSourceMalaria:Triazolopyrazine_%28TP%29_Series. Accession date: 2016 5 23.
-
Open source drug discovery – A limited tutorial.Parasitology141(1):148‑157. [InEnglish]. DOI: 10.1017/s0031182013001121
-
InChI in the wild: an assessment of InChIKey searching in Google.Journal of Cheminformatics5(1):10. [InEnglish]. DOI: 10.1186/1758-2946-5-10
-
Global Technical Strategy for Malaria 2016–2030.WHO Library Cataloguing-in-Publication Data,32pp. [InEnglish]. URL: http://www.who.int/malaria/publications/atoz/9789241564991/en/ [ISBN978 92 4 156499 1]
-
World Malaria Report 2015.WHO Library Cataloguing-in-Publication Data,280pp. [InEnglish]. URL: http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/ [ISBN978 92 4 156515 8]
Supplementary material
Download file (12.25 MB)