|
Research Ideas and Outcomes :
Guidelines
|
|
Corresponding author: Lyubomir Penev (l.penev@pensoft.net)
Received: 09 Nov 2022 | Published: 16 Nov 2022
© 2022 Donat Agosti, Laurence Benichou, Wouter Addink, Christos Arvanitidis, Terence Catapano, Guy Cochrane, Mathias Dillen, Markus Döring, Teodor Georgiev, Isabelle Gérard, Quentin Groom, Puneet Kishor, Andreas Kroh, Jiří Kvaček, Patricia Mergen, Daniel Mietchen, Joana Pauperio, Guido Sautter, Lyubomir Penev
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Agosti D, Benichou L, Addink W, Arvanitidis C, Catapano T, Cochrane G, Dillen M, Döring M, Georgiev T, Gérard I, Groom Q, Kishor P, Kroh A, Kvaček J, Mergen P, Mietchen D, Pauperio J, Sautter G, Penev L (2022) Recommendations for use of annotations and persistent identifiers in taxonomy and biodiversity publishing. Research Ideas and Outcomes 8: e97374. https://doi.org/10.3897/rio.8.e97374
|
|
Abstract
The paper summarises many years of discussions and experience of biodiversity publishers, organisations, research projects and individual researchers, and proposes recommendations for implementation of persistent identifiers for article metadata, structural elements (sections, subsections, figures, tables, references, supplementary materials and others) and data specific to biodiversity (taxonomic treatments, treatment citations, taxon names, material citations, gene sequences, specimens, scientific collections) in taxonomy and biodiversity publishing. The paper proposes best practices on how identifiers should be used in the different cases and on how they can be minted, cited, and expressed in the backend article XML to facilitate conversion to and further re-use of the article content as FAIR data. The paper also discusses several specific routes for post-publication re-use of semantically enhanced content through large biodiversity data aggregators such as the Global Biodiversity Information Facility (GBIF), the International Nucleotide Sequence Database Collaboration (INSDC) and others, and proposes specifications of both identifiers and XML tags to be used for that purpose. A summary table provides an account and overview of the recommendations. The guidelines are supported with examples from the existing publishing practices.
Keywords
semantic publishing, taxonomy publishing, semantic annotation, biodiversity, persistent identifiers, taxa, specimens, sequences, treatments, XML, JATS, TaxPub, tagging
Introduction
Specifics of taxonomic publications
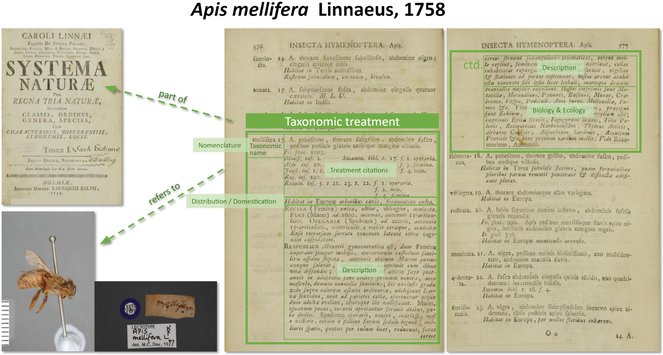
Taxonomic publications communicate the discovery of new biological taxa or new data on already known taxa in the form of taxonomic treatments, well delimited sections of text for each taxon (Fig.
Schematic representation of the taxonomic treatment of Apis mellifera Linnaeus, 1758. Sources: text: https://doi.org/10.5962/bhl.title.542; figures: https://linnean-online.org/16019/; composite: https://doi.org/10.5281/zenodo.5168465.
Translated into today’s digital world, this simple framework of presenting biological taxa in both human readable and machine interpretable format is sufficient, given that it is present as digital accessible knowledge (DAK,
Research results presented in the biodiversity literature are one of the best curated data (
In today’s digital arena these structured texts are an ideal prerequisite to enhance the publications by making them readable or even machine actionable (
Use of identifiers
An identifier (ID) is a label for any subject, conceptual, physical or digital (
As an identifier serves to unequivocally label an entity, it may also be employed to track the use of it, particularly when that entity is digital. Performance indicators are an important tool for the efficient management and development of organisations and infrastructures. Such indicators are used to channel appropriate funding internally, and also to request funding externally. The more we are able to show the impact and reach of our field, the easier it is to gather financial support to develop natural history collections, maintain services, digitise objects and conduct research.
Parallel to the rise of e-publishing, IDs minted and used in biodiversity informatics have diversified and become increasingly important to link to objects, specimens, and their digital representation, as well as the component parts of literature (
Usage of identifiers can be broad and complex. PIDs are used to identify and link digital and physical objects or concepts. One of the very first uses of DOIs was identifying individually published articles as well as references in the bibliographies, which enhanced the visibility and citability of these articles. DOIs are also used for data and figures, and are proposed for the digital objects in DiSSCo (
The Internet’s world wide coverage clearly makes it evident that globally unique identifiers are a prerequisite to locate the cited resources, and, consequently, through conversion and transformation of data, to build a knowledge graph, where all these resources can be identified and linked to each other through their PIDs. Since digitization of objects (e.g., an article) can occur in parallel, this can lead to collision between identifiers for physical objects, or across domains, between articles or specimens. Identifiers of different kinds have a long tradition in biodiversity research — they served specific purposes such as to label specimens from an expedition or a natural history collection, and have been understandable within their respective context. These identifiers served internal purpose and therefore only had the requirement to be locally unique. They are not resolvable through the internet making them hard to use by non-specialists or machines, as you will need to know the format to interpret them and there is no way to know if they are correct when used outside their source system. Assigning PIDs resolves these problems but introduces new challenges: the global uniqueness and opaque string requirements for persistence makes them hard to use for humans. Therefore additional IDs/labels for usage by humans, not necessarily globally unique, is also needed, which is not a problem as long as these are not used for data linkage. However, this will require look-up tables linking historic identifiers with the respective PIDs or extending non-unique IDs with a prefix to make it unique. Ideally the connection between the legacy ID and the unique PID is made either at the metadata level of each object, or within the specimen record (material citation) in publications.
Because of the current transitional period of digitising biodiversity data, new and different kinds of PIDs might be minted for the same object. To connect different PIDs for the same object we will need a discovery mechanism to build look-up tables. The different data accessible via the resolution of the PIDs will then provide complementary, sometimes conflicting data about the same objects (such as is discovered by GBIF’s clustering mechanism*
To minimise the costs of the significant and non-trivial effort of disambiguation of entities and building and maintaining look-up tables, the recommendations in this paper strongly encourage the use of harmonised PIDs that are compliant with a community accepted standard across different journals and publishers and serve, therefore, multiple scientific disciplines or domains. A good basis for harmonisation, for example, are the recommendations of the European Open Science Cloud (EOSC) for the use of PIDs that should be taken into account (
On the need of harmonisation
The recommendations in this paper are produced collaboratively by several organisations, research projects and biodiversity scientists. They are based on nearly 15 years of experience on annotating unstructured legacy publications by Plazi (
The paper has been largely elaborated and finalised in a collaboration between several partners in the Biodiversity Community Integrated Knowledge Library project (BiCIKL) (
Taxonomy is ruled by nomenclatural codes which state the requirements for a nomenclatural act to be validly published, whether in print or online. These rules have evolved with the emergence of online journals, and mandate the use of certain identifiers within the publication and especially in the full-text XML of articles, for example the LSID of the publication in which a new nomenclature act is published, or the mention of the ISSN for the journal (see
The objective of this paper is to list the main structural elements and data types present in taxonomic publications, the existing identifiers currently in use, and make proposals for use of additional PIDs where these do not exist yet. The paper aims at providing both recommendations, best practices and practical advice to technical editors and publishers in taxonomy on how to implement identifiers in their work and how they can be leveraged. For each element, the use of an identifier is discussed from the perspective of taxonomic publishing, its pros and cons are given, and short explanations of how and where to implement these PIDs. We recommend that authors and publishers provide as many identifiers and links as possible, facilitating in this way the conversion of the published content into a digitally accessible knowledge. This would be not only a starting point for the reuse of this important data at scale, but also spur new research based on this incredibly rich resource. It will also allow linking data in taxonomy with other scientific disciplines to build the future practice of evidence-based knowledge, that is to bridge the gap from a taxonomic name to machine actionable data about it.
Publications, publication sections, sub-article data elements and their identifiers
Modern taxonomic articles follow a rather strict structure that facilitates their representation in a structured XML format following the widely used TaxPub*
In its broader sense, publishing is the act of making content available to the public. In this paper, we refer specifically to peer-reviewed publications, either in the form of monographs (books) or periodicals (journals), print or electronic. While not all taxonomic publications are peer-reviewed, most of the comments and recommendations made here would apply to them too. Publishing taxonomic content, and specifically publishing nomenclatural acts, has a specific meaning and requires compliance to the rules, which are defined in various codes of nomenclature.
For Zoology, the International Code for Zoological Nomenclature (ICZN)*
publication, n.
- Any published work.
- The issuing of a work conforming to Articles 8 and 9.
electronic publication
A publication issued and distributed by means of electronic signals.
publish, v.
- To issue any publication.
- To issue a work that conforms to Article 8 and is not excluded by the provisions of Article 9.
- To make public in a work, conforming to (2) above, any names or nomenclatural acts or information affecting nomenclature.
In botany and mycology, the International Code of Nomenclature for algae, fungi, and plants (ICN)*
Publication is effected, under this Code, by distribution of printed matter (through sale, exchange, or gift) to the general public or at least to scientific institutions with generally accessible libraries. Publication is also effected by distribution on or after 1 January 2012 of electronic material in Portable Document Format (PDF; see also Art. 29.3 and Rec. 29A.1) in an online publication with an International Standard Serial Number (ISSN) or an International Standard Book Number (ISBN).
Front matter of the publication
Definition
The article’s front matter contains metadata for the article and its host journal: title, authors’ list with their affiliation, the date of publication, abstracts, keywords, a copyright statement, etc. (see front matter structures in JATS XML here*
Persistent unique identifiers for the front matter
International Standard Serial Number (ISSN)
Definition
The ISSN is an 8-digit number used to uniquely identify a serial publication. The system was designed in 1971, then published as a standard in 1975, and can be used for a journal as well as for book series, and even for some websites in the scholarly domain. It is unique and designates the publication medium, for instance if a journal is published in both print and digitally it must have a different ISSN for each media: a Print-ISSN and an E-ISSN (a different ISSN should also be given for any mobile version or CD-Rom version). One also needs an ISSN in case of a different language version of the same journal. When the publication is provided in different media, it is recommended to display all ISSN numbers on each version of the publication, if the latter is published, e.g. in different languages in different journals. The ISSN does not offer any resolution mechanism and is only a media-oriented identification.
Why does a journal need an ISSN?
The ISSN is mandatory for any journal or serial publication. In taxonomy, to be compliant with most nomenclatural codes, the nomenclatural acts should be published in a journal or series identified with an ISSN or a book with an ISBN.
Indeed, the International Code of Nomenclature for algae, fungi, and plants (ICN) stipulates that a nomenclatural novelty to be considered effectively published, it should be present in a publication distributed either in print (through sale, exchange, or gift) to the general public or at least to scientific institutions with generally accessible libraries, or (after 1 January 2012) in an online publication with an International Standard Serial Number (ISSN) or an International Standard Book Number (ISBN) and in Portable Document Format (PDF) (see also Art. 29.3 and Rec. 29A.1).
The International Code of Zoological Nomenclature (ICZN) stipulates in Art. 8.5 (
- have been issued after 2011
- have the date of publication stated in the work itself
- be registered in Zoobank (see below)
- contain the evidence of such registration (LSID of the publication or of the new name must be indicated in the work itself).
In Zoobank, the entry must have the name of an organisation other than the publisher that intends to permanently archive the work in a manner that preserves the content and layout, and is capable of doing so. The ISSN or ISBN of the publication must be registered in the Zoobank entry.
How to discover an existing ISSN
To find the ISSN of a series or journals, one may consult the ISSN portal, which provides a comprehensive list of ISSNs and some associated metadata.
How to obtain an ISSN
To get an ISSN for a journal or series, all the necessary information, is available at https://portal.issn.org/requesting-issn. In some countries the ISSN might not be free and may require a registration fee between 25 € and 50 €, depending on the country assigning the ISSN.
It seems possible to obtain an ISSN before the first publication of a print serial, however, it is very common to be asked to wait until number 2 of the series to be printed. Online publications are usually assigned an ISSN after the first or second issue is published (with at least 5 publications published), or in some countries, after the website of the new periodical has gone live and is fully functional.
How to annotate and display an ISSN
Display: ISSN: 1313-2989 (for the print version of a serial, if exists)
e-ISSN: 1313-2970 (for the online version of a serial, if exists)
For linked data purposes, it is even better to use the resolvable version of the ISSN: https://portal.issn.org/resource/ISSN/1313-2989
Annotate in JATS:
Tag the ISSN number using the <issn> element, using the publication-format attribute to specify the format or medium of the publication (e.g., “print”, “electronic”, “video”, “audio”, “ebook”, and “online-only”)*
<issn publication-format="ppub">[ISSN number]</issn>
<issn publication-format="epub">[ISSN number]</issn>
Example of an ISSN
2118-9773 is the ISSN of the European Journal of Taxonomy (EJT). As the journal is an e-only journal, it has only one online or e-ISSN.
ZooKeys has two ISSN depending on its version: 1313-2989 (for the print version) 1313-2970 (for the online version).
An ISSN can also identify a series of books, e.g. The Mémoires du Muséum national d’histoire naturelle have one for the print version (ISSN: 1243-4442) and one for the online version (e-ISSN: 1768-305X).
Sample annotation in JATS (for ZooKeys):
<journal-meta>
<issn publication-format="ppub">1313-2989</issn>
<issn publication-format="epub">1313-2970</issn>
</journal-meta>
Recommendation
Considering that ISSN (or ISBN) are mandatory for online publication in taxonomy to be compliant to both ICN and ICZN codes, and that an ISSN makes your journals or series more easily identifiable and findable, attributions of an ISSN or ISBN to taxonomic publications must be considered mandatory. A unique ISSN should be assigned to each version of the journal, print and electronic. Each linguistic version of the journal should also have its own ISSN.
International Standard Book Number (ISBN)
Definition
The ISBN was internationally approved as an ISO standard in 1970, and published in 1972, and is a unique international identifier for monographic publications. Correct use of the ISBN allows different product forms and editions of a book, whether printed or digital, to be clearly differentiated, ensuring that it identifies the specific version it relates to. Similarly to the ISSN, each version of the book, print, e-book, pdf etc., must have a different ISBN. A book included in a book series, or published as a monograph in a journal, can be provided with both an ISBN and the ISSN of the series in which it is published.
ISBN is a 13-digit number that identifies a book. As it is typically used in a barcode format, it is prefixed by an European Article Number (EAN). It is constructed as it shown in Fig.
Why does a book need an ISBN?
ISBN is important for cataloguing a book and for its findability, discovery, and dissemination. Its display is obligatory in the first pages of the book, along with the book title, author(s) name(s) and the publisher. ISBN is the main international record of your publication and is important for indexing and dissemination. It aims at facilitating the compilation of book trade directories and bibliographic databases, which in turn facilitate their dissemination as book dealers can use them to order books efficiently and unambiguously.
In taxonomy, it is crucial to have ISBN assigned to any taxonomic monograph with nomenclatural acts. For instance, as explained above, Zoobank requires an ISBN to register a nomenclatural act published within a book (
How to discover an existing ISBN
As a unique identifier, ISBN is part of the metadata associated with any book. To find the ISBN of any published book, whatever version of the book, PDF, e-book or print version, a simple query on the internet with the title followed by the mention of the ISBN will bring the answer. WorldCat is a good place to retrieve all the ISBNs of a book. Beware that a book may have as many ISBNs as format versions: one ISBN for the print version, another one for the ebook, or for second edition and so on.
How to obtain an ISBN
All the information needed to get an ISBN for a publication is available at https://www.isbn-international.org/content/how-get-isbn.
When an ISBN has been assigned to a publication, it should always be displayed to facilitate its identification. The ISBN is also crucial for dissemination as it is displayed in a barcode format, so libraries and bookshops can process incoming stock and outgoing sales quickly and accurately. On a printed book, an ISBN should be included on the copyright page, also called the title verso page, or at the foot of the title page if there is no room on the copyright page. If there is no barcode, then the ISBN should also be on the back cover or jacket preferably on the lower right. Each version of the book needs to be provided with its own ISBN. More details on when to assign an ISBN are available at https://www.isbn-international.org/content/isbn-assignment.
The publisher will then fill in the ISBN in the legal deposit form with all the additional metadata of the book for cataloguing purposes at their respective national ISBN agencies.
How to annotate and display an ISBN
Display: ISBN: 978-2-85653-939-2
Annotate in JATS:
Tag the ISBN number using the <isbn> element, using the publication-format attribute to specify the format or medium of the publication (e.g., “print”, “electronic”, “video”, “audio”, “ebook”, and “online-only”)*
<isbn publication-format="[format type]">[ISBN number]</isbn>Example of an ISBN
The Flora of New Caledonia published by the Muséum national d’Histoire naturelle in 2020 on Apocynaceae, Phellinaceae and Capparaceae has an ISBN for its print version (978-2-85653-939-2), one for the PDF version on sale (978-285653-954-5) and one for its bundle (print + e-book: 978-2-38036-955-2).
Sample annotation in JATS with multiple ISBN numbers:
<book-meta>
<isbn publication-format="print">978-2-85653-939-2</isbn>
<isbn publication-format="PDF">978-285653-954-5</isbn>
<isbn publication-format="bundle">978-2-38036-955-2</isbn>
<book-meta>Recommendation
An ISBN is mandatory to properly identify a published book. Each version of the book (PDF, print, ebook, each linguistic version, second edition) should have its own ISBN. Considering that ISSN and ISBN are mandatory for nomenclature purposes, we must consider the use of ISBN mandatory for taxonomic publications.
Digital Object Identifier (DOI)
Definition
The DOI system has been developed by the DOI Foundation and is implemented through a federation of registration agencies. The two most commonly used agencies that register DOIs in the scholarly domain are Crossref and DataCite. Both are membership organisations providing DOIs to research outputs but for different purposes. The main difference lies in the type of digital objects they identify, the scale of numbers of DOIs needed and the metadata associated with the DOI.
Crossref is a non-profit membership organisation specifically serving scholarly publications. Its members are publishers, research institutions, university presses, societies and funders. Membership in Crossref is open to organisations that produce professional and scholarly materials and content. In addition, applicants should be able to meet the terms and conditions of membership.
DataCite is a global non-profit organisation that provides persistent identifiers (DOIs specifically) for research data and other research outputs and resources. DataCite’s members work with data centres, stewards, libraries, archives, universities, publishers and research institutes that host repositories and who have responsibility for managing, holding, curating, and archiving data and other research outputs.
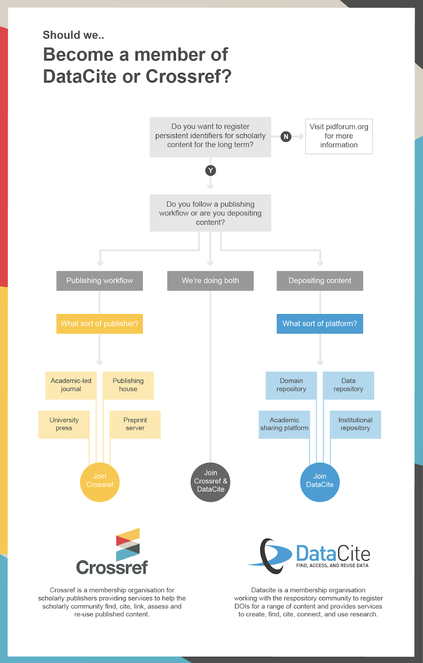
In their respective websites, a schema (Fig.
Dichotomic decision tree for the registration of CrossRef and DataCite DOIs, from the DataCite - Crossref release.
The DOI includes three parts Fig.
To create the DOI, the DOI prefix given to an organisation is combined with a suffix of choice. The DOI becomes active once registered with a DOI registration agency like CrossRef or DataCite. CrossRef provides a complete documentation on best practices to construct the suffixes.
How to discover an existing Digital Object Identifier (DOI)?
To find the corresponding DOI registered, enter the title, the author or any metadata in Crossref or DataCite search engines or use alternatively the ReFindit tool.
How to mint a Digital Object Identifier (DOI)
All agencies providing DOIs are listed here: https://www.doi.org/registration_agencies.html. Each of them may have different rules and apply different fees. Alternative repositories to mint DOI for legacy publications are the Biodiversity Heritage Library, the Biodiversity Literature Repository and institutional libraries retro-digitising legacy publications, such as E-Periodica at the Federal Institute of Technology, Zurich.
To deposit a DOI to Crossref,one has to be a member. Membership fees begin at 275 USD and depend on the revenue of the applicant. Once a member, a DOI prefix is assigned to the joining organisation and will form the stem of links to all its metadata records. Fees vary per record type, books, research grants, preprints, etc., from 0.15 USD for a legacy article to 1 USD for a newly published article. Each DOI has to be registered by direct deposit of XML, using Open Journal System Plugin for instance or, alternatively, through an online web deposit form.
Component DOIs are often registered for figures, tables, and supplemental materials associated with a journal article. They have their own metadata distinct from that of the parent article DOI.
The registration of the DOI includes all the metadata, i.e. basic information such as dates of publication, publication outlet, including the ISSN or ISBN, article title and authors. There is a Crossref membership obligation: accurate metadata should be deposited for all DOI registered, and the metadata should be maintained for the long term, including updating any URLs that change. It is also an obligation to include DOIs in the reference lists for existing works which have DOIs. A free public API is available to retrieve all existing Crossref DOIs.
To register a DOI with DataCite, one has to be a member. Membership is open to all organisations whose missions include research output sharing. A membership fee of 2,000 euros applies to member organisations. Once a member, non-for-profit members will have to pay another 500 € annual fee to make use of DOI registration services. Each DOI, up to 1,999, will cost 0,80 €. There are two ways to register a DOI: using an API or a Web Interface. All information is provided at https://support.datacite.org/docs/getting-started.
How to annotate and cite a DOI
Cite:
https://doi.org/10.3897/zookeys.1083.72939 (preferred), or DOI: 10.3897/zookeys.1083.72939
Annotate in JATS:
Tag the cited DOI with the <ext-link> element, using the xlink-href to provide the DOIs https version and the ext-link-type attribute with value “doi”.*
<ext-link xlink:href="[https-version of DOI]" ext-link-type="doi">[https-version of DOI]</ext-link>Example of a DOI
https://doi.org/10.3897/zookeys.1083.72939 refers to an article published in ZooKeys.
Sample annotation in JATS:
<ext-link xlink:href="https://10.3897/zookeys.1083.72939" ext-link-type="doi">
https://doi.org/10.3897/zookeys.1083.72939
</ext-link>Recommendation
Use Crossref DOI for articles and bibliographic references. For supplementary material, figures or tables, create Crossref component DOI or DataCite DOI. Generate a DataCite DOI for data. If none is available, try to find a way to create a DOI using an alternative repository such as BHL, BLR or E-Periodica, or DOIs issued for datasets deposited at large international repositories, such as GBIF, DataONE, Dryad, Zenodo and others.
Display all the identifiers, ISSN, ISBN, DOI, on the corresponding publication page and register all the corresponding metadata associated with the DOIs with CrossRef or DataCite. Always include the DOI in the metadata for other publication-related registration purposes, for example at ZooBank, IPNI, MycoBank, Zenodo, Dryad and others.
Body of the article
Definition
Most academic journals require the authors to write their articles following the IMRaD format. IMRaD stands for Introduction, Method, Result and Discussion which are the four main sections that constitute the structure of most scientific papers in the Science, Technical and Medical (STM) fields. The body of the article is the main textual and graphic content of the article and is situated between the front and the back matters. This usually consists of sections, subsections, and paragraphs, which may themselves contain figures, tables, etc.
In a taxonomic article, the body of the article includes specific items, such as taxonomic treatments, material citations, descriptions, differential diagnoses, details of collecting permits, etc.
Sections
Definition
Most journal articles are divided into sections, each with a title that describes the content of the section, such as “Introduction”, “Materials and Methods”, or “Conclusions”*
What are the identifiers for sections
Sections are normally tagged with internal Universally Unique Identifiers UUIDs in the article XML. In addition, the names of the sections, which are used more or less consistently in various science domains, e.g., “Introduction”, “Material and Methods”, “Results”, “Conclusions” etc. can be used for inferring a semantic meaning of their content, an approach that is currently used for the conversion to RDF and export to the OpenBioDiv knowledge graph.
How to annotate sections
In JATS, the sections are annotated using the following elements and attributes:
<sec sec-type="[section type]" id="[internal identifier]">
<sec-meta>
<mixed-citation>
<object-id object-id-type="uuid">[UUID]</object-id>
</mixed-citation>
...
</sec>The sec-type attribute annotates the basic structural unit of the body of a document. Following the recommendation that sec-type “is most useful when a list of values is maintained, and articles are tagged accordingly”, for JATS the values: "cases", "conclusions", "discussion", "intro", "materials", "methods", "results", "subjects", "supplementary-material", are recommended.*
The “id” attribute is a unique internal identifier of an element; it allows the element to be cross-referenced [and linked to]. The value must be unique across a document…[id] holds an internal document identifier that can be used by software to perform a simple link. An id should not be confused with elements that are used to hold externally defined identifiers such as a DOI”*
Though not recommended, a lighter-weight solution for associating an external identifier with a section is to “overload” the id attribute of <sec> by using an external identifier such as a UUID as the value. However, the “id” attribute “must start with a letter of the alphabet”*
Example
Annotation of a section “Methods” including an object identifier taken from the article of
<sec sec-type="methods" id="SECID0E1H">
<sec-meta>
<mixed-citation>
<object-id object-id-type="uuid">07ED460C-7F70-42EC-A7F8-59CC2F512131</object-id>
</mixed-citation>
</sec-meta>
<title>Methods</title>
<p>The specimens of the genus <em>Xiphocentron</em> studied here were borrowed
from the collections of the National Museum of Natural History, Smithsonian Institution in
Washington, DC, and from the Colección Nacional de Insectos, Instituto de Biología de la
Universidad Nacional Autónoma de México.</p>
<p>The type materials are deposited as indicated in each species description, in the
collections: National Museum of Natural History, Smithsonian</p>
</sec>Recommendation
Section and subsection titles should be tagged as such and Internal UUIDs should be assigned to them in the article XMLs.
Figures, figure captions and citations
Definition
A figure is either a photo or a scientific drawing illustrating biological species or part(s) of them, landscapes, habitats or equipment, or visualisation of data or results from statistical analyses. Figures and their captions convey an essential part of the information contained in a scientific paper and are of particular interest for the community.
The ICN states the importance of illustrations in its Art. 43.2:
“A name of a new fossil-genus or lower-ranked fossil-taxon published on or after 1 January 1912 is not validly published unless it is accompanied by an illustration or figure showing the essential characters or by a reference to a previously and effectively published such illustration or figure.”
According to article 40.3, illustrations can also be a type specimen prior to 1 January 2007*
The figures related to a taxonomic treatment (see definition below) are usually cited at the beginning of the treatment and are part of it.
What are the identifiers for figures
DOIs being either Crossref component DOIs or DataCite DOIs are usually used when the figures are deposited in a repository.
How to mint an identifier for a figure
For minting DOIs, see section “Digital Object Identifiers” above. If no DOIs are minted for figures, these can be identified with internal UUIDs minted by software during the compilation of the full-text article XML, and a hash of the figure allows to uniquely identify the respective figure.
How to annotate and cite a figure, figure caption and figure citation
Cite: Figures are cited within the text following the long-established practice in scholarly publishing (e.g., “according to Fig. X” or “see detail (Fig. Y)”). Citation style should follow the journal’s or publisher’s instructions for the authors.
Annotate in JATS:
Figure:
<fig id="[Internal identifier]">
<object-id object-id-type="doi">[DOI]</object-id>
<caption>[free_text]</caption>
</fig>In-text figure citation:
<xref ref-type="fig" rid="[Internal identifier]">[Figure reference]</xref>Note that the citation details of the figure as delivered by the Biodiversity Literature Repository (BLR) at Zenodo should contain both the DOI of the article and the component DOI of the figure. In the case where no CrossRef component DOI exists, a DataCite DOI is minted for the figure at BLR. The “rid” (reference to an identifier) attribute is needed to perform the linking with the <fig> element via the embedded “id” element.
Examples
Annotation of a figure and in-text figure citation (from
Figure:
<fig id="F4">
<object-id object-id-type="doi">10.3897/zookeys.1111.77586.figure1</object-id>
<caption>
<p>Malaise trap across Kaputu Stream at 1535 m altitude in the Mazumbai Forest Reserve in
the West Usambara Mountains, northeastern Tanzania (Photo: Trond Andersen).</p>
</caption>
</fig>In-text figure citation:
<xref ref-type="fig" rid="F4">Fig. 4</xref>https://doi.org/10.3897/zookeys.1088.78139.figure1 is a CrossRef component DOI assigned to Figure 1 of the article https://doi.org/10.3897/zookeys.1088.78139.
Recommendation
Use Crossref component DOI to identify each figure within an article. The component DOI has the important feature of a link from the figure DOI to its parent article DOI. If no Crossref DOI is available, use alternatives from DataCite.
In all cases, and especially if no DOIs are minted for figures, it is recommended to assign internal UUIDs minted by software during the compilation of the full-text article XML as well as a hash for unique identification.
When compiling the full-text XML, it is highly recommended to cross-reference (anchor) the in-text figure citations to their respective figures in the article body.
Tables, table citations
Definition
A table is a concise and effective way of presenting large amounts of data usually displayed in rows and columns for reference.
Tables are increasingly important because they contain, in many cases, a compilation of the specimens used, their sequence accession codes, specimen codes that allow linking to the cited specimens, as well as traits, such as measurements or qualitative descriptions or even the results of an analysis performed on the raw data taken from the specimens or from their environment. Each row can be envisioned to represent a structured material citation, and if used to list species used in a study, together with a taxonomic name, an entire taxonomic treatment.
What are the identifiers for table
In TreatmentBank, tables are identified by a UUID and a persistent http URI ID. In the Pensoft article XMLs, tables are identified by internal UUIDs.
How to mint a table identifier
DOIs or Crossref component DOIs relating to the article, should be minted and submitted for registration to Crossref by the publisher. If no DOIs are minted for tables, these can be identified with internal UUIDs minted by a software during the compilation of the full-text article XML.
Annotating and citing tables
Tables are cited within the text following the long-established practice in scholarly publishing (e.g., “according to Tab. X” or “see data (Tab. Y)”). Citation style should follow the journal’s or publisher’s instructions for the authors.
Annotate in JATS:
Table:
<table-wrap id="[Internal identifier]">
<object-id object-id-type="doi">[Digital Object Identifier]</object-id>
<caption>Text</caption>
<table>
...
</table>
</table-wrap>In-text table citation:
<xref ref-type="table" rid="[Internal identifier]">[Table reference]</xref>The “rid” attribute is needed to perform the linking to the <table> element via the “id” attribute of the target <table-wrap> element, which itself has optional, repeatable <object-id> elements recording identifiers for the table it contains. The JATS tag library defines the <object-id> element as a “Unique identifier (such as a DOI or URI) for a component within an article (for example, for a figure or a table)”, further stating that, “the <object-id> element holds an external identifier, typically assigned to an object such as a table by a publisher. The contents of this element should not be confused with the "@id" attribute, which holds an internal document identifier that can be used by software to perform a simple link inside the document.”*
Examples
Annotation of a table and in-text table citation (from
Table:
<table-wrap id="T1">
<object-id content-type="table" object-id-type="doi">
10.3897/zookeys.1111.77586.table1
</object-id>
<caption>Table 1. African genus Chimarra</caption>
<table>
...
</table>
</table-wrap>In-text citations:
<xref ref-type="table" rid="T1">Tab. 1</xref>Recommendation
Ideally, a table should be provided with a Crossref component DOI related to the article. In all cases, and especially if no DOIs are minted for tables, it is recommended to assign internal UUIDs minted by software during the compilation of the full-text article XML.
When compiling the full-text XML, it is highly recommended to cross-reference (anchor) the in-text table citations to their respective tables in the article body.
Taxonomic treatments
Definition
Taxonomic treatments are sections of publications documenting the features or distribution of a related group of organisms (taxon) (
The features and structure of treatments have changed over time, and vary between and within publications. Often an indication follows the name of whether the taxon is new to science, e.g., “species nova”, “sp. nov.” or “genus novum”, “gen. nov.” and the name or names of the persons who attribute the naming. A listing of taxa that are already known to science, citations of earlier treatments (treatment citations), often follows in a section. In cases when taxonomic names change as a result of a taxonomic revision, for example because of a raise in its rank, or because a taxon is synonymized, this is followed by a label stating the change, such as for example “syn. nov.” or “nov. stat.”. Other information, such as persistent identifiers and references to physical specimens, may also be included in a treatment.
A number of other sections may follow the nomenclature section. One of the most significant sections, frequently titled “Materials Examined”, includes citations to specimens used as the basis of the treatment and data about their properties (e.g., DNA sequences). This section often includes the circumstances of collection and/or deposition at a museum or other institution. Historically, these details have allowed scientists to visit the holding institution, or to seek a loan, for further scientific investigation of the same material that was described by the treatment. Also common is a “Description” section providing information — often in highly structured language, and sometimes in tabular form — on the distinctive features of the collected organisms, with an aim toward characterising the entire taxonomic class such material represents.
Similar to a “Description” section, there is a “Diagnosis” section, which contains descriptions of only those features or unique combinations of features “that distinguish that species from others, in the same way that the disease identification you receive when you visit the doctor is called the diagnosis because the doctor has distinguished your illness from all other possibilities based on the basis of your symptoms and tests” (
Similar to publications and following the FAIR principles, the treatments can be extracted from the publications, preserved separately and made freely accessible to the public (Fig.
An example of a treatment accessible in various formats. Source: https://europeanjournaloftaxonomy.eu/index.php/ejt/article/view/1639/5873
Alternative format:
TreatmentBank (TB) HTML: https://tb.plazi.org/GgServer/html/03E2AB75FFD6BE60FE39FCF5FBF9F83B
TB XML: https://tb.plazi.org/GgServer/xml/03E2AB75FFD6BE60FE39FCF5FBF9F83B
TB RDF: https://github.com/plazi/treatments-rdf/blob/main/data/03/E2/AB/03E2AB75FFD6BE60FE39FCF5FBF9F83B.ttl
An XML tagset for Taxonomic treatments has been formalised as an extension of the Journal Article Tag Suite (JATS) (
Treatments are reused by GBIF upon extraction, where they are imported as part of a dataset in a Darwin Core Archive format compiled from taxonomic treatments and cited figures. Currently these article-based datasets represent almost 60% of all the datasets published in GBIF.
In Wikidata, taxonomic treatments can be annotated with the property taxonomic treatment (P10594)*
The Barcode of Life Data Systems (BOLD) Barcode Identification Numbers (BINs) (
The UNITE Species Hypotheses (SHs) are functionally similar to treatments including all the clustered public fungal ITS sequences to which a unique DOI is assigned by UNITE. UNITE is a database and sequence management environment for the molecular identification primarily of fungi but now also of other taxa. It focuses on nuclear ribosomal internal transcribed spacer (ITS) region sequences that are considered the fungal barcode. All species hypotheses have a unique URL where the associated public sequences are displayed (
What are the identifiers for taxonomic treatments
DOI
A subtype “taxonomictreatment” has been added in Zenodo as a DataCite digital object identifier (DOI) to the “publication” type. The metadata for taxonomic treatments in Zenodo are enhanced with added custom keywords based on existing domain specific vocabularies (e.g., Darwin Core), links to the source publication, cited figures or related identifiers such as the http URIs minted by TreatmentBank (see below). In case of treatments deposited in BLR via TreatmentBank, the respective HttpURI are included in the metadata.
For BINs*
HTTP URI
The “HttpURIs” were created by Plazi for treatments in 2009 parallel to the development of the persistent HTTP URIs for specimens now widely accepted in CETAF. The HTTP URIs are used by GBIF when reusing TreatmentBank treatments. The HTTP URIs are kept persistent and are built based on a unique UUID and the prefix “http://treatment.plazi.org/id/UUID” (e.g., http://treatment.plazi.org/id/0000C505-BB5D-484C-76BE-9AB6999DEB23). The original intention was to share the UUID with Zoobank whereby the Zoobank UUID would resolve to the taxonomic name and to the respective taxonomic treatment in TreatmentBank. Unfortunately this synchronisation has been discontinued.
UUID
During the publication of a taxonomic article, Pensoft journals assign UUIDs to each taxon treatment. Those UUIDs are further used by Plazi to mint the HTTP URIs of the treatments at TreatmentBank.
How to discover treatment identifiers
The DOI of a treatment can be found by searching ReFindit or for those minted by Biodiversity Literature Repository, through the search engines of Zenodo or TreatmentBank. The HTTP URIs can be found through GBIF or the Biodiversity Literature Repository (BLR) or TreatmentBank.
Via ReFindit API (search by author, year, and taxon name, the latter as title):
https://refindit.org/find?search=advanced&author=Kronestedt&year=2011&title=Pardosa%20zyuzini (requires some further matching to pick correct result)
Via Zenodo UI (full text search):
https://zenodo.org/search?page=1&size=20&q=Pardosa+zyuzini+Kronestedt+2011
Via TreatmentBank statistics API (exact match search on author, year, and taxon name):
Via TreatmentBank UI search (fuzzy search for taxon name):
Via GBIF UI (dataset search):
https://www.gbif.org/search?q=Prosymna+lisima
Via GBIF API (species search):
How to mint an identifier for treatment
Currently, Zenodo is the only place to mint a DOI for a treatment. UUIDs are generated by some publishers during the article processing before publication (all Pensoft journals, for example). HTTPURIs are minted by TreatmentBank. This is not an exclusive solution, however, since a treatment is a subtype of the DataCite publication type at Zenodo.
How to annotate and cite treatments
Cite: A citation of a treatment can be provided either by its DOI or its HTTP URI generated by Plazi’s TreatmentBank. The citation of other treatments normally happens within a given treatment’s Nomenclature section (in the so-called “nomenclature-citation-list” of the JATS/TaxPub XML representation), where they can also introduce a nomenclatural change, indicated with a label (e.g. syn. nov., comb. nov., nom. nov., etc.).
Annotate in JATS (treatment and subsections in JATS/TaxPub):
<tp:taxon-treatment>
<tp:treatment-meta>
<mixed-citation>
<object-id object-id-type="doi">[digital object identifier]</object-id>
</mixed-citation>
</tp:treatment-meta>
</tp:taxon-treatment>For the nomenclature subsection:
<tp:nomenclature>
<tp:taxon-name>[Taxon name]</tp:taxon-name>
</tp:nomenclature>For all other subsections:
<tp:treatment-sec sec-type="[section type name]"> ... </tp:treatment-sec>Section types should, if possible, make use of the following vocabulary terms: description, diagnosis, discussion, distribution, ecology_behavior, conservation, etymology, materials_examined, reference_group, and vernacular_names which will add a semantic meaning to (sub-)section titles and facilitate the extraction and reuse of the data.
Examples
Annotation of treatments, nomenclature section and subsections:
Treatment:
<tp:treatment-meta>
<mixed-citation>
<object-id content-type="doi">10.5281/zenodo.6964440</object-id>
</mixed-citation>Nomenclature section:
<tp:nomenclature>
<tp:taxon-name>Tribasodites yatung</tp:taxon-name>
</tp:nomenclature>Subsection:
<tp:treatment-sec sec-type=”description"> ... </tp:treatment-sec>Recommendation
Tag each taxonomic treatment in the article full-text XML and then assign a CrossRef Component DOI or Datacite DOI or internal UUID for it. Register all the metadata associated with the DOI.
Treatment citations
Definition
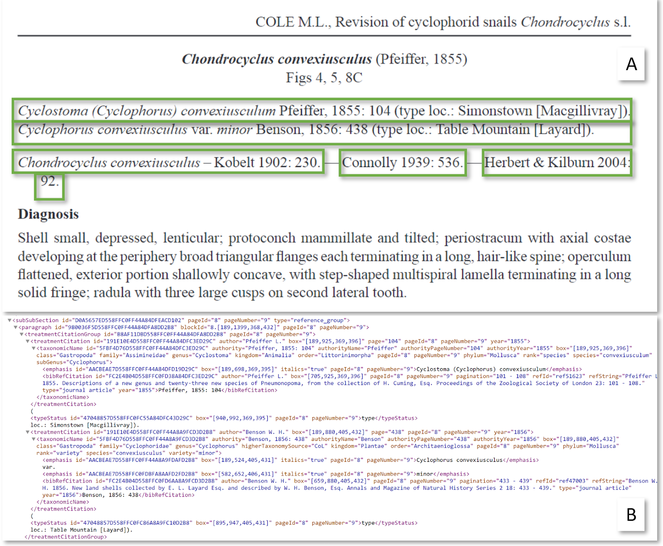
A treatment citation is a reference to a previous treatment, in many cases the original description of the taxon, or protologue (Fig.
Treatment citation. Green boxes: treatment citations. A. published example. B: annotated treatment citations in TreatmentBank using TB internal XML.
Source: https://europeanjournaloftaxonomy.eu/index.php/ejt/article/view/787/1829
TreatmentBank XML: https://treatment.plazi.org/GgServer/xml/101687E3D558FFC3FDF9A837FECED008
Treatment citations are the source and basis for creating synonymic lists and taxonomic catalogues.
Treatment citations are analogous to bibliographic references in a publication citing previous works.
What are the identifiers for treatment citations
No identifiers are known, however, citations can and should be tagged in the backend XML of the article to be made discoverable and processed for further use.
How to discover treatment citations
Treatment citations are listed subsequent to the nomenclatural sections of a taxonomic treatment. They usually consist of a taxonomic name, the authority and year, and a page number, especially in zoology. In combination, the authority and year are also a bibliographic citation of the original publication of the respective treatment, albeit often implicit, because traditionally, taxonomists do not include this kind of bibliographic references in the article reference list (
How to mint an identifier for treatment citation
There is no established procedure for minting treatment citations, except for possible assignment of internal UUIDs to them.
How to annotate and cite a treatment citation
The treatment citation annotations are attributed with persistent HTTP URIs of the respective treatment(s) in TreatmentBank. The treatment citation element is currently being remodelled and thus the recommendations might change in the next version of TaxPub.
Annotate in JATS/TaxPub:
<tp:nomenclature>
<tp:taxon-name>
<tp:taxon-name-part taxon-name-part-type="genus">Cus</tp:taxon-name-part>
<tp:taxon-name-part taxon-name-part-type="species">dus</tp:taxon-name-part>
</tp:taxon-name>
<tp:nomenclature-citation-list>
<tp:nomenclature-citation>
<tp:taxon-name>Aus bus</tp:taxon-name>
<mixed-citation>
<object-id content-type="taxonomic_treatment" object-id-type="doi">[DOI]</object-id>
</mixed-citation>
</tp:nomenclature-citation>
</tp:nomenclature-citation-list>
</tp:nomenclature>Example
Annotation of treatment citation in the treatment of Chondrocyclus convexiusculus (Pfeiffer, 1855) in
<tp:nomenclature-citation-list>
<tp:nomenclature-citation>
<tp:taxon-name>Cyclostoma (Cyclophorus) convexiusculum</tp:taxon-name> Pfeiffer, 1855: 104
(Type loc.: Simonstown [Macgillivray]).
</tp:nomenclature-citation>
<tp:nomenclature-citation>
<tp:taxon-name>Cyclophorus convexiusculus var. minor</tp:taxon-name> Benson, 1856: 438
(type loc.: Table Mountain [Layard]).
</tp:nomenclature-citation>
</tp:nomenclature-citation-list>
<tp:nomenclature-citation-list>
<tp:taxon-name>Chondrocyclus convexiusculus</tp:taxon-name> –
<tp:nomenclature-citation>Kobelt 1902: 230</tp:nomenclature-citation>
<tp:nomenclature-citation>Connolly 1939:536</tp:nomenclature-citation>
<tp:nomenclature-citation>Herbert & Kilburn 2004: 92</tp:nomenclature-citation>
</tp:nomenclature-citation-list>Recommendation
Treatments should be cited by their PIDs, either through their inclusion in a nomenclature-citation in a nomenclature section of the citing treatment or as a standalone in-text citation in any part of the article as follows: “Based on Treatment: [hyperlinked treatment PID, where a treatment PID can be either the DOI of the treatment provided by BLR or Plazi’s HTTP identifier available from TreatmentBank] I conclude that ....”.
Treatment citations should be tagged in the article XML as separate entities and, if available, should contain the existing PIDs of the cited treatments.
Recently, a joint statement of CETAF, SPNHC and BHL has been published (
-
Provide each scientific name of a taxon, at least on its first mention in the paper, with authorship, date, and corresponding entries to the publication’s “Bibliographic references” section.
-
If the publisher’s guidelines do not allow you to list it as a reference, cite it properly as a bibliographic reference by adding the page number after the date for instance. For example, for a species described in EJT http://dx.doi.org/10.5852/ejt.2022.828.1851 on p. 48, it is preferable to use the notation Infrantenna fissilis Liu & Sittichaya, 2022: 48 instead of Infrantenna fissilis Liu & Sittichaya, 2022.
-
Provide the corresponding persistent identifier (PID) to each of these references where they exist, i.e. a Crossref DOI minted by the publisher or minted by the Biodiversity Heritage Library (BHL) when the legacy publication has been digitised retrospectively and provided with a DOI, or a DataCite DOI minted by organisations digitising legacy literature (e.g., e-Periodica at the Federal Institute of Technology Zurich) or the Biodiversity Literature Repository (BLR) at Zenodo.
-
Provide the PID of the taxonomic treatment where they exist, using for instance, the DOI of the treatment deposited in BLR, or for articles with primary taxonomic descriptions minted by BHL (for example: https://www.biodiversitylibrary.org/part/304567).
Material citations
Definition
A material citation is a reference to, or citation of, one or multiple specimens in scholarly publications (https://dwc.tdwg.org/terms/#materialcitation;
The GBIF occurrences can create a rich linking network for specimens because a GBIF specimen record can be linked to a material citation published in a scholarly article, or at least to the treatment or publication containing that record.
What are the identifiers for material citations
TreatmentBank and the Biodiversity Data Journal issue internal UUIDs for material citations. They are reused in conjunction with the treatment UUID in GBIF in the form of “treatment UUID.mc.material citations UUID”. GBIF is minting an identifier for each material citation present as an occurrence record in their infrastructure. TB maintains the links and identifiers of the occurrences in GBIF with their respective material citations in TreatmentBank.
How to discover material citations' identifiers
These identifiers are currently minted post-publication by TreatmentBank, or before publication by the Biodiversity Data Journal, and can be found using TreatmentBank data access interface (https://tb.plazi.org/GgServer/srsStats) which can also provide access to the related GBIF occurrence ID.
Via TreatmentBank statistics UI with a wide variety of search fields and output fields to choose from. The link (visit TreatmentBank statistics UI) shows taxon name, author, year, and type status.
Via TreatmentBank statistics API with a wide variety of search fields and output fields to choose from in the UI. The link (visit TreatmentBank statistics API) shows taxon name, author, year, and type status, retrieved as JSON. A more simplified API is under development.
Via GBIF API (occurrence search for taxon name, restricted to materials citations).
http://api.gbif.org/v1/occurrence/search?basisOfRecord=MATERIAL_CITATION&scientificName=Lebertia+insignis Other search fields are also available, e.g. country, might require further matching efforts to find additional matches from specific source publications in GBIF.
How to mint an identifier for material citation
Follow your standard procedure for minting UUIDs.
How to annotate and cite material citation
Annotate in JATS/TaxPub:
Use “object-id” to provide an identifier for a material citation in the article which allows it to be cited unambiguously.
<tp:material-citation>
<object-id content-type="[content type]">[Identifier]</object-id>
[material citation string]
</tp:material-citation>To provide an external identifier for a component of a material citation (e.g., a catalog number or occurrence id), use <named-content>, specifying the type of identifier in the content-type attribute.
<named-content content-type="[content type]">[Identifier]</named-content>The <uri> element may be used to tag an identifier that is a URI and provide a live link to the representation of the identified resource:
<named-content content-type="[content type]">
<uri xlink:href="[URI]">[URI]</uri>
</named-content>Examples
A material citation from Monomorium dryhimi that can be unambiguously cited (
<tp:material-citation>
<object-id content-type="arpha">B5596AA1-CDF9-DDA3-D5CD-D922E1723751</object-id>
Holotype worker. SAUDI ARABIA, Al Bahah province, Amadan forest, Al Mandaq
governorate, 20°12'N, 41°13'E, 1881 m.a.s.l. 19.V.2010 (M. R. Sharaf &
A. S. Aldawood Leg. KSMA
</tp:material-citation>A material citation citing a specimen from the MNHN Paris (
<tp:material-citation>
<named-content content-type="dwc:occurrenceID">
<uri xlink:href="http://coldb.mnhn.fr/catalognumber/mnhn/p/p04158076">
http://coldb.mnhn.fr/catalognumber/mnhn/p/p04158076
</uri>
</named-content>
…
</tp:material-citation>Finer grained markup
<tp:material-citation>
<object-id content-type="arpha">B5596AA1-CDF9-DDA3-D5CD-D922E1723751</object-id>
<tp:type-status>Holotype</tp:type-status> worker.
<tp:material-location>King Saud Museum of Arthropods (KSMA),
College of Food and Agriculture Sciences, King Saud University, Riyadh,
Kingdom of Saudi Arabia.
</tp:material-location>
<tp:collecting-event>
<tp:collecting-location>
<tp:location>
SAUDI ARABIA, Al Bahah province, Amadan forest, Al Mandaq governorate
</tp:location>
</tp:collecting-location>,
<named-content content-type="dwc:verbatimCoordinates">20°12'N, 41°13'E</named-content>
, 1881 m.a.s.l. 19.V.2010 (
<named-content content-type="dwc:recordedBy">M. R. Sharaf</named-content>
&
<named-content content-type="dwc:recordedBy">A. S. Aldawood</named-content>
Leg.);
</tp:collecting-event>
</tp:material-citation>Besides GBIF issuing an occurrence ID for the material citations, and Pensoft’s Biodiversity Data Journal, no other publishers are using IDs for material citation so far. For EJT and the journals of the MNHN Paris, Plazi is adding the material citations attribute after extracting the data from the published papers.
In legacy publication annotations, material citations are attributed with a unique UUID in TreatmentBank. These UUIDs are resolvable via Plazi SRS*
The TreatmentBank UUID for the material citation is reused in GBIF as a couple of treatment UUID * material citation UUID:
Identifier = 03A10B47FFDFFFAFFDE0FA60FB18F865.mc.3B60B00CFFD1FFAFFF38FED7FD4AFE22
In the Biodiversity Data Journal, the material citations are exported to Darwin Core Archive and indexed by GBIF automatically on the date of publication. The internal material citation UUID is minted and entered in the “occurrenceID” of Darwin Core. If the “occurrenceID” is already occupied by the original ID supplied by the author, it should be moved to the “associatedOccurrences” field of Darwin Core, while the “occurrenceID” field should be used again for the internal material citation ID provided by the journal.
Example from Biodiversity Data Journal:
<tp:treatment-sec sec-type="materials
<list list-type="alpha-lower" list-content="occurrences">
<list-item>
<p>
<bold>Type status:</bold>
<named-content content-type="dwc:typeStatus">Holotype</named-content>.
<bold>Occurrence:</bold>
catalogNumber:
<named-content content-type="dwc:catalogNumber">
<ext-link xlink:href="[url]">IFRD9449</ext-link>
</named-content>;
recordedBy:
<named-content content-type="dwc:recordedBy">Liu Yu-Wei</named-content>;
occurrenceID:
<named-content content-type="dwc:occurrenceID">UUID</named-content>;
associatedOccurrences:
<named-content content-type="dwc:associatedOccurrences">
living culture IFRDCC3104
</named-content>
</p>
</list-item>
</list>
</tp:treatment-sec>Recommendation
Publishers should use unambiguous separators, such as a Unicode character U+2022 “•”, for the material citations within an article and identify these with UUIDs in the backend article JATS XML. When material citations represent a holotype or other type specimens, this specific status, the collecting event and the collection should be tagged unambiguously in the backend XML to facilitate harvesting and reuse.
Taxonomic names
Definition
A taxonomic name, or more generally scientific name, is the formal name, that is the scientific identity, given to a species or, more generally, a taxon, following the rules of nomenclature and used widely beyond taxonomy to link data to a particular taxon. Although the concept of scientific names, along with rules on the interrelationships of taxa, was introduced in the ancient times by
Identifiers for new taxa descriptions and other types of nomenclatural acts, and their online registration, are used increasingly, and the process is regulated by zoological (ICZN) and botanical (ICN) codes (
The Catalogue of Life (COL) consortium, in a collaboration with the Global Biodiversity Information Facility (GBIF), aims to provide a global list of accepted names (
The National Centre for Biotechnology Information (NCBI) taxonomy database holds unique identifiers (taxIDs) for taxonomic names for which sequence data is available at the INSDC (
What are the identifiers for new names or nomenclatural acts
Fungi
Pre-publication registration of identifiers for names, typifications and other nomenclatural acts is mandatory for fungi since 1st January 2013. The identifiers must be published in the protologue or in nomenclatural changes.
Living vascular plants: IPNI (International Plant Names Index)
In botany, the registration of nomenclatural acts was accepted at the XIX International Botanical Congress in Shenzhen 2017 (
Post-publication indexing is a well-established practice of the IPNI which covers seed plants, ferns and lycophytes, but not bryophytes or algae. IPNI is produced collaboratively by The Royal Botanic Gardens, Kew, The Harvard University Herbaria, and The Australian National Herbarium and is hosted by the Royal Botanic Gardens, Kew. Pre-publication indexing and inclusion of IPNI record identifiers in the publication was first implemented by the Pensoft journal PhytoKeys (
Algae
PhycoBank is the registration system for nomenclatural acts (new names, new combinations and types) of algae (
Fossil plants (except for fossil fungi and diatoms)
Pre-publication indexing is established in the Fossil Plant Names Registry (FPNR) and the International Fossil Plant Names Index (IFPNI). Registration of taxa is not mandatory.
Bryophytes
IDs for new bryophyte names can be obtained from the Index of Mosses Database (W³MOST).
Animals
ZooBank provides registration of new nomenclatural acts, published works, and authors. It is an authoritative online, open-access, community-generated registry for zoological nomenclature provided as a service to taxonomists, biologists, and the global biodiversity informatics community. It is also the official register of the International Commission on Zoological Nomenclature (ICZN).
The registration of Type Specimens is allowed in Zoobank but yet not fully implemented. Registration is mandatory for electronic publications publishing new nomenclatural acts since 1st January 2012. Each electronic publication receives an identifier (LSID) minted by ZooBank.
Identifiers for taxa in Catalogue of Life, NCBI taxonomy, and TreatmentBank
The Catalogue of Life and the NCBI taxonomy are two widely used reference taxonomies. Both issue taxon name IDs. For references in articles, authors can use hyperlinked taxon IDs of either COL or NCBI just as they use sequence accession numbers.
TreatmentBank mints persistent identifiers for taxonomic names as part of the annotation and FAIRizing of treatments in legacy literature. They are a combination of the treatment UUID extended with “.taxon”.
How to discover identifiers of names
The following web sites provide the search facility for discovering the identifiers of names.
Fungi
Living vascular plants
International Plant Names Index (IPNI)
Algae
Fossil plants (except fossil fungi and diatoms)
IFPNI (International fossil plant names index)
PFNR (Plant Fossil Names Registry)
Animals
Identifiers of nomenclatural acts can also be found through other services, for example the World Register of Marine Species www.marinespecies.org.
Catalogue of Life
https://www.checklistbank.org/tools/name-match
Via Catalogue of Life UI (advanced name search):
https://www.catalogueoflife.org/data/search?q=Pardosa+zyuzini
Via Catalogue of Life API (name usage search):
https://api.catalogueoflife.org/dataset/3LR/nameusage/search?q=Pardosa+zyuzini
Via GBIF UI (species search):
https://www.gbif.org/species/search?q=Pardosa+zyuzini (results include list of other identifiers)
Via GBIF API (species search):
https://api.gbif.org/v1/species?name=Pardosa+zyuzini (requires further matching to pick desired identifiers, e.g. ZooBank UUID of name)
NCBI taxonomy
https://www.ncbi.nlm.nih.gov/taxonomy
TreatmentBank
Identifiers for taxonomic names can be found using TreatmentBanks stats https://tb.plazi.org/GgServer/srsStats
How to mint an identifier for new names
Fungi
Mycobank is an on-line database aimed as a service for the mycological and scientific community by documenting mycological nomenclatural novelties, that is, new names and combinations, and associated data such as descriptions and illustrations.
Index Fungorum, the global fungal nomenclator coordinated and supported by the Index Fungorum Partnership, contains names of fungi including yeasts, lichens, chromistan fungal analogues, protozoan fungal analogues and fossil forms, at all ranks. As a result of changes to the ICN relating to registration of names, Index Fungorum provides a mechanism to register names of new taxa, new names, new combinations and new typifications.
Authors of novel fungal taxa must register the new names in only one registry, e.g. either in MycoBank or Index Fungorum or Fungal Names. These registries regularly coordinate sharing of data and have arranged an informal agreement to only accept the first listed name in case it appears in more than one registry. Registration of the same new name in multiple registries is considered an inappropriate practice that creates a considerable amount of confusion and extra work for the registries and necessitates the deprecation of the duplicated registrations at a later stage.
Living vascular plants
IPNI(International Plant Names Index)uses LSIDs as unique identifiers for plant names and provides a mechanism to register those LSIDs. IPNI records LSIDs for names of new taxa, new combinations and replacement namesfor living and vascular plants. LSIDs are not mandatory for valid publication of a plant name. However, if an IPNI LSID is needed, it can be pre-registered on the IPNI website. For new taxa, the holotype data can also be provided. The new plant name will be provided with a LSID that will be activated once the article is published. It is important to note that IPNI can only provide LSIDs for “vascular plants”, i.e., extant ferns, lycophytes and seed-bearing plants. Thus, IPNI will not give LSIDs for fungi, bryophytes (mosses), macroalgae (Rhodophyceae etc.), diatoms, or any fossil vascular plant.
Algae
PhycoBank is the registration system for nomenclatural acts such as new names, new combinations and types of algae (
Fossil plants (except fossil fungi and diatoms)
PFNR (Plant Fossil Names Registry) is a database of preferably new names, but also previously published names of plant fossils and associated nomenclatural acts excluding fossil diatoms and fossil fungi. It is run by the National Museum Prague for the International Organisation of Palaeobotany. A LSID links the name to its original publication. The registration of a new nomenclatural act results in a registration number that is added to the manuscript. This part is not public and, if necessary, all data can be changed during manuscript processing. These data are available only to the account owner who registered the manuscript, and to the editors of the database. When the paper is published, the missing data should be added and completed. A more detailed guide for name and typification registration is available.
IFPNI (International fossil plant names index) is a comprehensive literature-based record of the scientific names of all fossil plants, algae, fungi, allied prokaryotic forms, protists (ambiregnal taxa) and microproblematica. IFPNI provides an authoritative online, open-access, community-sourced registry of fossil plant nomenclature as a service to the global scientific community. A dynamic database documents all nomenclatural novelties including new scientific names of extinct organisms and associated data, including registration of the scientific publications containing nomenclatural acts and author-generated taxonomic literature in palaeobotany and palaeontology. IFPNI issues LSIDs for each kind of data object to locate biologically significant data over a network. LSIDs are designed to be automatically machine resolvable. Read more about IFPNI coverage.
Animals
To obtain a LSID for a new publication or a new name, the article has to be pre-registered in Zoobank by filling in a form with all the metadata: type of publication, article or monograph in a series, date of publication, authors, full title, ISSN of the journal, DOI of the article, volume, number, pages, online archive (
How to annotate and cite a taxon name
In JATS/TaxPub
<object-id object-id-type="Taxon name service">[taxonomic name identifier]</object-id>Examples of annotations
Fungi
The new fungal species Neopestalotiopsis rhapidis Qi Yang & Yong Wang bis, sp. nov., published in Biodiversity Data Journal (
In the article JATS XML, this record is annotated as:
<tp:taxon-treatment>
<tp:nomenclature>
<tp:taxon-name>
<object-id content-type="arpha">1EC88425-ADBF-5527-B845-9EF7658D0BA9</object-id>
<tp:taxon-name-part taxon-name-part-type="genus">Neopestalotiopsis</tp:taxon-name-part>
<tp:taxon-name-part taxon-name-part-type="species">rhapidis</tp:taxon-name-part>
<object-id object-id-type="MycoBank">840065</object-id>
</tp:taxon-name>
<tp:taxon-authority>Qi Yang & Yong Wang bis</tp:taxon-authority>
<tp:taxon-status>sp. nov.</tp:taxon-status>
</tp:nomenclature>
</tp:taxon-treatment>Living vascular plants
The new plant speciesArdisia whitmorei Julius & Utteridge, sp. nov. published in PhytoKeys (
In the article JATS XML, this record is annotated as:
<tp:taxon-treatment>
<tp:nomenclature>
<tp:taxon-name>
<object-id content-type="arpha">7A38F1AF-338B-5EAE-BE17-7FE4DD689BD9</object-id>
<object-id content-type="ipni">urn:lsid:ipni.org:names:77302868-1</object-id>
<tp:taxon-name-part taxon-name-part-type="genus" reg="Ardisia">
Ardisia
</tp:taxon-name-part>
<tp:taxon-name-part taxon-name-part-type="species" reg="whitmorei">
whitmorei
</tp:taxon-name-part>
</tp:taxon-name>
</tp:nomenclature>
</tp:taxon-treatment>Algae
The new algae Halamphora kenderoviana Zidarova, P.Ivanov, Dzhembekova, M.de Haan & Van de Vijver, published in PhytoKeys (
In the JATS XML of the article, this record is annotated as:
<tp:taxon-treatment>
<tp:nomenclature>
<tp:taxon-name>
<object-id content-type="arpha">55756560-6844-5EE2-9343-68965B35BA9C</object-id>
<named-content content-type="phycobank" xlink:href="https://www.phycobank.org/103140">
Phycobank 103140
</named-content>
<tp:taxon-name-part taxon-name-part-type="genus" reg="Halamphora">
Halamphora
</tp:taxon-name-part>
<tp:taxon-name-part taxon-name-part-type="species" reg="kenderoviana">
kenderoviana
</tp:taxon-name-part>
</tp:taxon-name>
<tp:taxon-authority>
Zidarova, P.Ivanov, Dzhembekova, M.de Haan & Van de Vijver
</tp:taxon-authority>
<tp:taxon-status>sp. nov.</tp:taxon-status>
</tp:nomenclature>
</tp:taxon-treatment>Animals
A new species of giant Eunice, Eunice dharastii published in
In the article, the JATS XML is annotated as follows:
<tp:nomenclature>
<tp:taxon-name>
<object-id content-type="arpha" object-id-type="UUDI">
BFC45050-4831-5B2D-9E7C-F99DC97D5DF1
</object-id>
<object-id content-type="zoobank" object-id-type="UUID"
xlink:href="https://zoobank.org/63BC2367-9654-45DA-8021-FD17584DFFDC">
63BC2367-9654-45DA-8021-FD17584DFFDC
</object-id>
<tp:taxon-name-part taxon-name-part-type="genus" reg="Eunice">
Eunice
</tp:taxon-name-part>
<tp:taxon-name-part taxon-name-part-type="species" reg="dharastii">
dharastii
</tp:taxon-name-part>
</tp:taxon-name>
<tp:taxon-status>sp. nov.</tp:taxon-status>
</tp:nomenclature>Expression of links to taxon names in JATS from the Catalogue of Life and NCBI Taxonomy
The link to Formica rufa in the Catalogue of Life is as follows:
<object-id content-type="COL">https://www.catalogueoflife.org/data/taxon/6JGM9</object-id>The link to Formica rufa taken from the NCBI taxonomy
<object-id content-type="NCBI">
https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=258706
</object-id>Links to other taxon specific catalogues can be added by entering a respective content-type.
Linking a taxonomic name anywhere in the text to the taxonomic name in the respective treatment, an id and <xref rid> element can be included.
<tp:taxon-treatment>
<tp:nomenclature>
<tp:taxon-name id="tn1">Formic rufa
<object-id object-id-type="UUID">C41B87B7-C473-EE75-A7E8-8565FCE1A945.taxon</object-id>
</tp:taxon-name>
</tp:nomenclature>
...
</tp:taxon-treatment>In a following section of the same article
<sec>
<title>Conclusions</title>
<p>
In this study,
<xref rid="tn1">
<tp:taxon-name>
Formica rufa
<object-id content-type="UUID">C41B87B7-C473-EE75-A7E8-8565FCE1A945.taxon</object-id>
</tp:taxon-name>
</xref>
is different from Formica rufa sensu Collingwood, 1966.
</p>
</sec>
Recommendation
Provide a pre-publication registration and include identifiers of new taxa or nomenclatural acts in the original article whenever possible, even when this is not required by a Code. Where and how to register new taxa and get identifiers for different groups of organisms such as algae, fungi, plants, or animals is explained in the sections above.
Specimens
Definition
Physical specimens held in collections may be cited directly, for example, material citations as part of taxonomic treatments, or in other sections of the article. In other cases, data derived from the specimens such as genetic sequences may include a reference to the specimen source. To keep track of the use of these specimens, collections should assign them with at least locally, but better with globally unique IDs (catalogue numbers)*
Specimens are often cited by combinations of metadata other than the DwC triplet, such as a who-what-when-where combination, e.g. a specimen “X”, collected in locality “Y” by collector “Z” on date XX-YY-ZZZZ, belonging to Taxon A, identified by Person “B”. This may include names of the person(s) who collected it, where and when this happened and/or a taxonomic identification. A field number may also be used, which acts as a unique identifier for the collection event as minted by the collectors. These numbers are not unique beyond this narrow context and may not have a systematic syntax. The combination of these properties may allow a specimen to be uniquely identified, but this is not a trivial task and natural language processing as well as disambiguation efforts are required.
Increasingly, the aim is to keep track of physical specimens through digital twins, called Digital or Extended Specimens (
What are the identifiers for specimens
Darwin Core “triplet” of Institution Code, Collection Code and Catalogue Number
The Darwin Core triplet is a concatenation of three Darwin Core properties associated with the physical specimen:
-
institutionCode: a code that is commonly associated with the institution where the specimen is held. Often, this is the acronym or one of the acronyms of this institution’s name, either in English or in the native language. In botany, codes from the Index Herbariorum*
28 are commonly used. -
collectionCode: a code that describes a collection held at the institution indicated above. Institutions may curate multiple collections at different locations and/or with different underlying themes, such as higher taxonomy or geography.
-
catalogNumber: a (mostly) alphanumeric code, often a barcode, that is used by the curator of the specimen to uniquely identify it.
By combining institutional provenance and locally used identifiers, the triplet is a simple solution to turn specimen metadata into a globally unique identifier. While it is opaque to the specimen’s data, such as the who-what-when-where properties for the event when it was collected, it is constructed of human-readable metadata elements that do not necessarily require a resolver. Triplets have been adopted for this reason to different extent by various infrastructures and their data providers. At GBIF, for example, providers of specimen data still regularly publish data using triplets as occurrence IDs. The infrastructure itself uses the triplet as a fallback measure to keep track of updates to occurrence records for specimens, if the combination of the ID for the data provider and the occurrence ID is not sufficient. At INSDC, guidelines*
Triplets are concatenations that function unambiguously as machine-readable identifiers only if they are concatenated consistently using the same methodology, which is not always the case. In practice, the three elements may be separated by different characters, including “:”, “/”, “<” and “-”. The institution and collection elements may change, as institutional branding or internal organisation evolve. The use of simple acronyms and other codes carries the risk of introducing homonymous triplets. Also, since triples are not resolvable, typing mistakes in the triplets are not easily discovered and there is no way to know if the catalogNumber in the triplet actually exist. It has proven that Darwin Core Triplets are often riddled with semantic and syntactic errors (
Catalogue or Specimen Number
CETAF stable HTTP Identifiers
The CETAF specimen identifier concept was established to create a general PID for physical specimens in CETAF collections, rooted in the concept of Semantic Web*
CETAF IDs are globally unique by virtue of the Domain Name System, but require institutional investment and policy to ensure their persistence, as domain names may change and they need to be opaque to any potential technical modifications to the infrastructure hosting them. Failure to accommodate these requirements may lead to link rot. Additionally, CETAF IDs have the disadvantage of not being easily discoverable.
Digital Specimen PIDs
The Digital Specimen concept was coined in 2019 during the Biodiversity Next meeting in Leiden and will represent “a digitised physical specimen, containing information about a single specimen with links to related supplementary information” (
Physical object ID
The PhysicalObject is a DataCite type referring to an inanimate, three-dimensional object or substance used for artefacts or specimens*
How to discover specimen identifiers
How to mint a specimen identifier
Physical specimen identifiers such as a catalogue number or registration number should be linked to the physical specimens. For preserved specimens, this is often done through barcodes like QR-codes. They need to be minted by the curating institution.
CETAF identifiers are globally unique identifiers for the physical specimens and are also minted by the institutions, but require an IT infrastructure that implements the specification, and an institutional pledge to keep the URIs resolving even if the underlying infrastructure is changed.
Digital Specimen PIDs (DOIs) are likely to be minted by regional infrastructures responsible for maintaining the digital specimen infrastructure, such as DiSSCo.
Physical Object PIDs (DOI) can be minted using the Zenodo repository, IGSN, or DataCite.
How to annotate and cite specimens
Specimen IDs should be cited either through their inclusion in the specimen record, as with material citations, or as a standalone in-text citation as follows: “Based on Specimen: [hyperlinked physical specimen PID or digital specimen DOI, where a physical specimen PID can be any resolvable PID including e.g. CETAF ID, ARK, DOI], I conclude that….”, in tabular format, or alternatively, in supplementary material. The latter, however, is not preferred because of technical limitations to find and extract the data subsequently.
Annotate in JATS:
<named-content
content-type="issuing institution specimen identifier"
xlink:href="specimen http URI">
institution ID
</named-content>Examples
In the article of
In the article XML, this annotation is expressed as follows:
<named-content content-type="cetaf_specimen_id" xlink:href="https://uri_prefix/CETAF_ID">
CETAF_ID
</named-content>Recommendation
Use specimen identifiers whenever possible, especially when they are persistent and resolvable; introduce the practice of citing a specimen, analogously to INSDC accession numbers; keep the IDs separate from HTTPs in the backend XML; use Digital Specimens DOIs when they become available; authors should be encouraged to cite specimens through their IDs.
Whenever CETAF identifiers are available, authors should use them to cite specimens rather than combinations of variable specimen properties.
In case resolvable identifiers such as Digital Specimen DOIs or CETAF identifiers are not available, we recommend using the local catalogue number of a specimen with explicit mention of the Collection and/or Institutional Codes where the specimen is preserved. If resolvable identifiers are available for the collection or organisation (e.g. ROR or WikiData), use these instead of Collection or Institutional Codes.
Sequence data
Definition
Nucleotide sequence data has become fundamental in both basic and applied areas of research related to biology and living organisms. This includes DNA and RNA sequences, genomes and transcriptomes with optional annotations, metagenomes and raw sequence data among other data types.
Sequence data is usually submitted by researchers to public sequence repositories, such as the International Nucleotide Sequence Database Collaboration (INSDC) and cited in publications through their accession numbers in the INSDC databases such as GenBank, ENA, and DDBJ (see below for details). The sequence data is synchronised between the databases using the same accession number but with different prefixes. In ENA, the human readable access is by using the prefix https://www.ebi.ac.uk/ena/browser/view/ and the machine operable version by https://www.ebi.ac.uk/ena/browser/api/embl/. In NCBI it is https://www.ncbi.nlm.nih.gov/nuccore/, and in DDBJ it is http://getentry.ddbj.nig.ac.jp/getentry/na/".
What are the identifiers for gene sequences
INSDC accession numbers
The International Nucleotide Sequence Database Collaboration (INSDC) is a global initiative committed to sharing sequence data and its associated metadata. This collaboration includes three nodes, the DNA Data Bank of Japan (DDBJ), the European Nucleotide Archive (ENA) at EBI and GenBank at NCBI, that comprise the largest repositories of nucleotide sequence data.
INSDC accession numbers are unique identifiers assigned to data submitted to INSDC. These are unique and stable alphanumeric codes that identify each sequence, sample or project, and that also provide information about the type of data and the INSDC partner to which it was submitted*
BOLD Process IDs
The Barcode of Life Database (BOLD) is a platform developed at the Centre for Biodiversity Genomics in Canada for the storage and analysis of barcode sequence data. BOLD Process IDs are unique identifiers in the BOLD system, created to connect specimen metadata such as taxonomy, collection information and images, to the DNA barcode sequence. These have a standard format that includes the project code and a numeric code, followed by the year the record was submitted. The sequence ID corresponds to the Process ID followed by the genetic marker code sequenced.
All public sequences of the BOLD database are periodically mirrored to GenBank, so the public BOLD sequence IDs are associated with INSDC accession numbers.
How to discover gene sequences’ identifiers
Sequence data identifiers can be searched for in the respective sequence databases, but these are also linked to other specific molecular databases or portals and even associated with Operational Taxonomic Units (OTUs) such as BOLD Barcode Identification Numbers (BINs), or UNITE Species Hypothesis (SHs), or specimen and distribution data such as occurrences in GBIF (
Via ENA UI (search by taxon name):
https://www.ebi.ac.uk/ena/browser/text-search?query=Formica+selysi
How to mint an identifier for gene sequences
Accession numbers and other sequence identifiers are automatically generated when the data is submitted to the public databases.
How to annotate and cite gene sequences identifiers
Cite: Sequences, BINs or SHs should be cited either through their hyperlinked persistent identifiers included in the specimen record, that is, the material citation, or in other parts of the articles such as figure legends, tables, appendices or free text, as a standalone in-text citation in the following way, e.g.: “Based on Sequence [hyperlinked accession number], I conclude that….”
Annotate in JATS XML:
<named-content content-type="Institution Name" xlink:href="httpURI">accession name</named-content>Examples
In the main text:
“The following sequences [hyperlinked accession numbers] were used in the analysis… ”
In the Data accessibility section:
“DNA sequences: GenBank accessions xxx to yyy.”
“The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEBxxxx (https://www.ebi.ac.uk/ena/browser/view/PRJEBxxxx).”*
Sample annotation in JATS for the sequence COI: AM 932788:
NCBI (National Centre for Biotechnology Information)
<named-content content-type="NCBI" xlink:href="https://www.ncbi.nlm.nih.gov/nuccore/AM932788">
AM932788
</named-content>ENA (European Nucleotide Archive)
<named-content content-type="ENA" xlink:href="https://www.ebi.ac.uk/ena/browser/view/AM932788">
AM932788
</named-content>Recommendation
Publishers should take care not only to include accession numbers in the content but also hyperlink them to the source database and tag them in the backend article XML.
Annotation to only one institution, either ENA or NCBI, should be provided for each accession number.
Persons
Definition
People have different roles in publications, which can often be inferred. The role is identified by the context in which the person’s name occurs, which itself is indicated by annotating the section of text within which it occurs. A person appearing in the author section is an author of the publication. A person appearing after a taxonomic name has two roles: as authority of the taxonomic name and also possibly an author of the publication in which the taxon is described. A person whose name appears in a material citation is most likely either a collector or identifier of the cited specimen. A person’s name appearing in a short or long bibliographic reference is an author or editor of the cited publication. A person’s name in the etymology section is probably honorific, in which case at least part of the name will be Latinized in the taxon name (Article 60.8,
There are local and thematic biographical databases of scientists and collectors that provide lists of names, affiliations, birth and death dates and the period a scientist has been active (floruit). In many cases, locally unique identifiers are provided. Such repositories either have only the names of nationals or internationals affiliated to national institutions, while others also refer to non-locals, for example if they are co-authors or collaborate within projects.
Zoobank and IPNI provide LSIDs respectively for zoologists and botanists. For botany the International Plant Name Index (IPNI) also provides a standard form for taxonomic name authorities abbreviations widely used in publishing.
What are the identifiers for person names
ORCID
The Open Research and Contributor ID (ORCID) is an identifier for researchers, with the principal aim of uniquely identifying and connecting them to their publications. It is maintained by the not-for-profit ORCID organisation, which operates through fees paid by member organisations. These organisations are mainly research institutions such as universities and commercial publishers, all of which benefit from widespread ORCID adoption and can make use of the ORCID APIs. ORCID identifiers are a subset of International Standard Name IDs (ISNI), which extend beyond research to other media content creators.
ORCID identifiers have widespread adoption and support, and are easy to register and manage by the researchers themselves. They do suffer from a few downsides: ORCID profiles with scarce metadata and limited or no linked publications are difficult to disambiguate or track back to the person. This may happen when research institutions mandate ORCIDs for their staff, but these new records are only poorly maintained if at all and duplicates may even be created. ORCIDs are also not suitable for deceased researchers, as they are intended to be self-maintained. Finally, ORCID identifiers are intended for use by individuals, not groups, teams or organisations. Also, an individual can register several ORCIDs which adds considerable ambiguity. A particular downside of ORCID is the condition for confirming that only part of the personal data can be made public; despite that this is a GDPR-required condition, it makes the use of the data difficult in some cases. Still, ORCID identifiers are currently the most commonly used means to identify scientific researchers in publications.
ISNI (International Standard Name Identifier)
ISNI is an ISO standard (International Standard Name Identifier, ISO standard 27729) established in 2010, and is widely used to identify people as well as organisations involved in creative activities, and public personas of both such as pseudonyms, stage names, record labels or publishing imprints. The original ISNI database has been populated and is regularly updated from the Virtual International Authority File (VIAF) database. Thus, it is not used exclusively for people. An analysis of a random sample of 10,000 Wikidata items with an ISNI number reveals that about 90% are individuals, and thus ISNI identifiers seem to be widely used*
ISNI is an open standard and its database is populated by harvesting the information from other resources using matching algorithms. The ISNI community would like to promote its usage worldwide, but had to meet the challenges linked to the requirements of the Global Data Protection Regulations (GDPR). A revised version of their data policy was published in March 2021*
VIAF
The Virtual International Authority File is a service maintained by the cooperative Online Computer Library Centre (OCLC), a global organisation of libraries. In VIAF, multiple national authority files are compiled into a single authority file where authors are disambiguated. VIAF identifiers may be more appealing for use in taxonomic publications than ISNI identifiers as they can be used to identify authors through their work available in library catalogues, and because of their deliberate overlap (see ISNI).
Wikidata ID
Wikidata is an open graph database hosted by the Wikimedia Foundation. Similar to how Wikipedia was conceived as a community-curated encyclopaedia of all knowledge that is sufficiently notable, Wikidata is the same for linked data. Any Wikidata-notable*
Taxonomic researchers databases
Numerous databases exist to keep track of taxonomic names, treatments and literature. Many of these have data on people involved in taxonomic research and in observing/collecting specimens, for which person identifiers may be minted. These databases are often under closed curation, but the identifiers may be used to identify people who do not appear in any other system. For instance, LSIDs for authors are used by Zoobank for zoology and the International Plant Names Index (IPNI) for botany. Examples of such databases are also the List of the entomologists of the world on Wikipedia or the Harvard index of botanists.
ResearcherID
The ResearcherID is an identifier for authors, reviewers and editors of scientific publications. It is hosted and maintained by Clarivate, a commercial company that is also responsible for the Web of Science publication index, Endnote bibliography management software, and the Publons review tracking database. As such, this identifier easily connects authors to their work tracked by the Clarivate infrastructures, but paid access to these services is required*
Scopus Author ID
Scopus is the database of research publications maintained by the Dutch commercial company Elsevier. Author IDs are automatically minted as content enters this database, and may be merged or split as needed or prompted by author feedback forms. The system encourages authors to connect their Scopus Author ID to their ORCID profile, as they can manage the latter themselves*
How to discover persons’ identifiers
ORCID
ORCID IDs can be found through: https://orcid.org/orcid-search/search?searchQuery=name
ISNI (International Standard Name Identifier)
There is a database search engine to search ISNI identifiers: https://isni.org/page/search-database/
Wikidata IDs
Wikidata provides several ways – ranging from generic to very specific – to find identifiers for people. There is a generic search box available on every Wikidata page, and it can be used for searching with name strings (example). This often yields large numbers of results, including many irrelevant ones, so the query can be refined, e.g., to yield only humans (example) or humans meeting some additional criteria, be it an additional string like “botanist” (example) or an additional identifier like the ZooBank author ID (example) or any additional statement, e.g., a specific place of birth (example).
ResearcherID
Clarivate explains how to search an author identifier in the specific page in the Web of science core collection. However, one has to be registered to access the database.
Scopus Author ID
Use Scopus search engine at: https://www.scopus.com/freelookup/form/author.uri?zone=TopNavBar&origin=NO%20ORIGIN%20DEFINED
How to mint an identifier for person's name
ORCID
In the ORCID model, researchers are invited to register an ID for themselves, and include this ID whenever they publish new work. Many publishers are already ORCID members and technically support easy inclusion of ORCID with author metadata. The combination of this linked body of work and a few pieces of metadata, such as name and (past) affiliation(s), allows unique identification of researchers and facilitates keeping track of their interests, performance and collaborations.
ISNI
The website does not indicate the membership fees upfront. If you want to get an ISNI for yourself, you need to contact the registration agency that provides this service.
Wikidata
Wikidata is an open database, so it is very straightforward to add new records or amend existing ones. Any volunteer can contribute anonymously, in which case IP address will be logged, or through a free registered account. New content needs to comply with community guidelines or may be removed by other volunteers, and moderators can enforce stronger restrictions.
ResearcherID
Researchers can mint their own identifier by registering at https://www.researcherid.com/#rid-for-researchers. ResearcherIDs may also be minted automatically for researchers in the Clarivate system that appear in multiple records but have no ID yet.
Scopus Author ID
Scopus Authors profile’s are automatically generated by metadata extracted from documents indexed in Scopus. The profile cannot be edited by the researcher. If correction is required, a request has to be sent to Scopus. Scopus Authors IDs are aligned with ORCID.
How to annotate and cite person's name identifier
Cite: A person ID is usually cited only in the authors’ section.
Annotate in JATS:
<contrib contrib-type="author" corresp="no">
<contrib-id contrib-id-type="orcid">https://orcid.org/0000-0002-0241-1036</contrib-id>
<name name-style="western">
<surname>Liu</surname>
<given-names>Yuxin</given-names>
</name>
<xref ref-type="aff" rid="A1">1</xref>
</contrib>Examples
ORCID
0000-0001-5864-8676 is Philippe Bouchet’s ORCID
ISNI (International Standard Name Identifier)
0000 0001 2127 4957 is Carl Linnaeus’ ISNI
Wikidata
Q1043 is Linnaeus’ Wikidata ID.
Recommendation
For persons, ORCIDs are the recommended identifiers if available. If not, ISNI, VIAF, IPNI or Zoobank identifiers could be used if these do exist, which may be possible for people not involved in scientific research or who died before they could create their own ORCID. For any other case, Wikidata is the recommended resource. IPNI or Zoobank identifiers should be added for nomenclature purposes, if available.
Institutions and collections
Definition
The institution is an organisation or infrastructure having custody of the objects included in its holding. The collection or dataset can include specimens of a shared origin, history or collecting campaign, normally part of the activities of an institution. Collection is sometimes also used in the sense of institution, or one institution may have several thematic collections (e.g., Vertebrates, Insects, Non-insect invertebrates), thus often causing confusions between the two terms.
What are the identifiers for institutions
ROR
ROR is in many ways an equivalent of ORCID for research organisations. It is built on the data of the Global Research Identifier Database (GRID) system, which has a similar scope. ROR has replaced GRID as identifier for research organisations, GRID does not have public updates anymore and is curated by a commercial company. Like ORCID, ROR aims for an open approach to creation and maintenance of data, operating under community oversight and establishing close links to other infrastructures such as DataCite and CrossRef (
To identify research organisations, ROR is currently the most recommended option. However, many research institutions have hierarchical structures, with many different faculties, departments, labs, groups, libraries and archives branching off of a single organisation tree. ROR is intended only for the top-level institution, not any subunit. For legacy reasons, such subunits may still be present in ROR (and GRID) but this is under continuous discussion. Child-level organisations are only included in ROR if there are sufficiently independent from the top-level organisation, as for affiliations the linkages should preferably be done at the top-level. An example of an allowed child organisation with its own ROR is national museum that is part of a university. A department or lab would not get its own ROR. for organisational subunits there is no globally adopted identifier system yet. Other solutions should be found if identification of a lower-level subunit such as a department is needed. However, in the scope of persistent identifiers in taxonomy and biodiversity publishing these should be avoided.
GRSciColl
The Global Registry of Scientific Collections (GRSciColl) is a community-curated clearing house of scientific collections*
As a clearing house, GRSciColl does not currently provide a PID on its own, but is able to associate the following identifiers with entities, allowing GRSciColl to act as a collaborative space to link records. GRSciColl collections and GRSciColl institutions have a unique URL as part of the GBIF registry, however these are not guaranteed to be persistent and should therefore not be used as identifier (Table
| Institutions | GRSciColl ID, institutionCode* |
| Collections | GRSciColl ID, collectionCode* |
| Staff | ORCID ID, Wikidata* |
GRSciColl provides an open API*
NCBI Biocollections
The NCBI Biocollections database holds curated metadata for institutions and collections, e.g., natural history museums, culture collections or herbaria, associated with sequence records available at the INSDC. It is maintained by the NCBI taxonomy group and used to support the construction of the DwC triplet voucher annotations added to sequence and sample records at the INSDC (
How to discover an institution’s identifier
ROR
Query after name of the institution at: https://ror.org/search?query=name
GrSciColl
Use the search interface at: https://www.gbif.org/grscicoll/institution/search
NCBI
Search the list of biocollection at: https://www.ncbi.nlm.nih.gov/biocollections/advanced
How to mint an identifier for an institution
ROR
ROR works on a community-based curation model and creating a new ROR record or updating an existing one cannot be done directly. Any change can be proposed through a feedback form, after which it will be subjected to community discussion on a Github repository.
GrSciColl
As a community-based curated database, anyone can suggest updates to a GRSciColl record that will be submitted for approval to reviewers that may include institution editors, country mediators, or administrators. GBIF has implemented community-curation functionality, enabling those working within each of the institutions and collections, to help maintain up-to-date information in the registry.
NCBI
To register a new collection, send an email to the NCBI contact.
How to annotate and cite institutions' identifiers
Cite: An institution can be cited by its abbreviated name, provided it is fully spelled during the first use in the article and linked to a PID to prevent ambiguity.
Annotate in JATS:
<named-content
content-type="institution"
xlink:href="https://www.gbif.org/grscicoll/institution/09d260e0-a509-40c7-bef5-b0f5444be84a"
id="NCID0E3CAE">
Western Australian Museum
</named-content>
<aff id="A1">
<label>1</label>
<addr-line content-type="verbatim">Pensoft Publishers, Sofia, Bulgaria</addr-line>
<institution>Pensoft Publishers</institution>
<addr-line content-type="city">Sofia</addr-line>
<country>Bulgaria</country>
<uri content-type="ROR">https://ror.org/01znaqx63</uri>
</aff>Examples
The French National Museum of Natural History (MNHN, Paris) has 27 collections identified in GRSciColl. The same institution has a ROR Id (https://ror.org/03wkt5x30) (former GRID grid.410350.3), and several other IDs such as ISNI (0000000121749334), Crossref Funder ID 501100007522 and Wikidata Q838691.
The institution code for the Western Australian Museum
<named-content
content-type="institution"
xlink:href="https://www.gbif.org/grscicoll/institution/09d260e0-a509-40c7-bef5-b0f5444be84a"
id="NCID0E3CAE">
Western Australian Museum
</named-content>The institution code for Pensoft Publishers
<aff id="A1">
<label>1</label>
<addr-line content-type="verbatim">Pensoft Publishers, Sofia, Bulgaria</addr-line>
<institution>Pensoft Publishers</institution>
<addr-line content-type="city">Sofia</addr-line>
<country>Bulgaria</country>
<uri content-type="ROR">https://ror.org/01znaqx63</uri>
</aff>The institution code for Naturalis, Leiden
<named-content
content-type="dwc:institutionCode"
xlink:title="National Museum of Natural History, Naturalis"
xlink:href="http://grbio.org/institution/national-museum-natural-history-naturalis">
RMNH
</named-content>Recommendations
Encourage institutions to include their ROR in GRSciColl and in GBIF datasets.
Use ROR for author’s affiliations and specimen affiliations, or another PID such as Wikidata ID or GRSciColl for specimens if a ROR is not available.
Encourage institutions to ensure their metadata is up to date in GRSciColl. Ensure that the collectionCode and institutionCode used on specimen records match codes registered in GRSciColl.
Make use of the Darwin Core terms collectionID and institutionID on specimen records, populated with the PID that is included in a GRSCiColl collection or GRSCiColl institution record.
Promote the use of GRSciColl PIDs as a means to link entities.
Ensure that all identifiers related to an institution are linked one to another, e.g., make sure all identifiers of the institution are mentioned in its ROR ID.
Back matter
Definition
The article back matter contains information that is ancillary to the main text, such as cited references, acknowledgements, declaration on authors’ contributions, declarations on conflicts of interest, declarations on funding, footnotes, supplementary materials, appendices, glossary, etc. The JATS structure for back matter is listed here.
This kind of information can be included either in the article text, or in the article back matter metadata alone, or in both places which may often cause confusion.
Acknowledgements
Definition
Section at the end of the manuscript where the authors thank colleagues or institutions who helped them in their work, e.g., contributed to it in producing some research, provided information, assisted the research, reviewed the manuscript, etc. It also mentions and acknowledges the projects and grants that funded the research. Linking funding agencies, grant numbers and persons to their PIDs or their homepages is potentially a very important source of information to build alternative metrics for individual contributions or the impact of a funding agency.
What are the identifiers in the ackowledgement
Persons
For persons contributing to the publication, use the <contrib> tag.
Funding agencies
Funding agencies are the institutions funding research including science foundations, other funding agencies, private charity trusts, or others. Funding agencies normally provide a grant award number that needs to be cited. There are, however, two large international sources that list projects and funders with their identifiers, the EU infrastructure OpenAIRE and the Funder Registry at CrossRef.
How to discover funding agencies identifiers
Both OpenAIRE and Crossref Funder Registry provide a search interface and API which helps publishers integrate the data about projects and funders with their editorial systems. Wikidata has different properties to acknowledge funding or for general acknowledgements.*
How to annotate and cite identifiers from the acknowledgment
Funding agencies
A funding agency should be cited by its name, tagged and linked with its identifier to avoid ambiguity and promote findability. The following example includes the agency name, the grant award number as well as a DOI of the agency name.
Person names
See the respective section above.
Examples
- Funder: European Commission. Funded project: Building the European Biodiversity Observation Network (EU BON). Grant number: 501100000780.
- Funding program: Institute of Museum and Library Services, award # LG-246400-OLS-20, "Extending data curation to interdisciplinary and highly collaborative research" (from Kouper and Cook KJ, 2021, https://doi.org/10.3897/biss.5.79084).
Annotate in JATS:
<funding-source>
<institution-wrap>
<institution content-type="funder_name">XXX</institution>
<institution-id institution-id-type="funder_doi">XXXX</institution-id>
<institution-id institution-id-type="funder_identifier">XXX</institution-id>
</institution-wrap>
</funding-source>Sample annotations in JATS:
<funding-source>
<institution-wrap>
<named-content content-type="project_title">
EU BON: Building the European Biodiversity Observation Network
</named-content>
<named-content content-type="project_identifier">308454</named-content>
<named-content content-type="project_funder_id">501100000780</named-content>
<named-content content-type="project_funder_name">European Commission</named-content>
<named-content content-type="project_funder_doi">
http://doi.org/10.13039/501100000780
</named-content>
</institution-wrap>
</funding-source>
<funding-source>
<institution-wrap>
<named-content content-type="funder_name">
Institute of Museum and Library Services
</named-content>
<named-content content-type="funder_identifier">100000208</named-content>
<named-content content-type="funder_doi">http://doi.org/10.13039/100000208</named-content>
</institution-wrap>
</funding-source>Recommendation
The Acknowledgment (as well as the List of references sections) should be included in the manuscript text.
Authors' contributions, supporting funders/projects and declarations of conflict of interest should be part of the machine-readable article back matter metadata. A common and highly recommended way to facilitate the conversion to machine-readable format is to request this information during the submission process through APIs or dropdown list of items for: (1) CrossRef Funder Registry and OpenAIRE databases, from which the authors can select the acknowledged funder and research project; (2) CRedIT taxonomy*
Funder and project names in the backend article XML should be assigned external identifiers, such as Funders’ IDs from OpenAIRE or CrossRef Funder Registry and grant numbers, when acknowledging grants.
We recommend authors to fill in supporting grants and funders also in the article metadata during the submission process. The integration of OpenAIRE and CrossRef’s Funder Registry during the submission system would disambiguate the grant and funder titles/names which should be included in the article-meta tag of JATS.
Bibliographic references and citations
Definition
The references cited within a paper are listed at the end of the manuscript and usually organised by alphabetical order based on the surname of the first author, and often include the DOI of the publication. They refer to the sources used and quoted in the text. In the humanities, the bibliography often refers to all the materials consulted and sources the authors went through to conceive their research, and are not all necessarily cited in the text. On the contrary, in STM (Science, Technical and Medicine fields), all references relate to in-text citations. All in-text citations must refer to a reference listed at the end of the article.
Bibliographic citations have a specific use in taxonomy. Indeed, traditionally a taxonomic name is followed by the mention of the author who first described it. A bibliographic citation is implicit in the taxonomic name, and both together, the taxon name and the authority, form the “taxon concept”. For example, “L. 1758” in Formica rufa L. 1758 is a bibliographic reference to Linnaeus, 1758. Bénichou et al. 2018 recommend adding a proper bibliographic reference, such as Formica rufa, Linnaeus, 1758: 580, and then add the full bibliographic reference in the list of references. This will help facilitate the reader to find the referenced article. From a machine actionable point of view, it will enable building a citation network of the article, and give credit to the authors of the referenced publications. Other publishers such as Pensoft and Magnolia Press have been recommending this for a long time, however it has not been mandatory because of the additional burden on the authors for compiling excessively long reference lists for all taxon names mentioned in a paper.
Another specific use of a citation that has yet to gain wider traction is to indicate the citation context, for which CiTO – the Citation Typing Ontology*
What are the identifiers for bibliographic references
Digital Object Identifiers
DOIs issued by CrossRef by or on behalf of the publisher are most commonly accepted as a norm by most publishers. BHL provides Crossref DOIs for digitised articles from legacy literature. BLR at Zenodo provides DataCite DOIs for legacy articles which do not have DOIs or for several sub-article elements such as treatments, figures and others.
Handle
Handles are sometimes used by institutional repositories such as the Digital Repository at the American Museum of Natural History (AMNH), however, their use in the publishing world is very limited.
How to discover bibliographic references identifiers
The DOIs can be found using the search form provided by CrossRef or DataCite for their minted DOIs, or Refindit, which includes all the providers of DOIs.
How to mint DOIs
DOIs are minted by the publishers, or on behalf of publishers, and subsequently submitted to CrossRef for registration. DataCite DOIs are minted by the DataCite partnering organisations and can be taken from there by the data aggregators and publishers.
CrossRef DOIs are preferred since their metadata fit perfectly their bibliographic purpose. For legacy publications, one may also check repositories such as BLR at Zenodo that provide free DataCite DOIs so to retrieve the exiting DOI.
How to annotate and cite them
The DOI follows the bibliographic reference and is hyperlinked (see example below)
Annotate in JATS:
<ext-link
ext-link-type="doi"
xlink:href="https://10.5962/bhl.title.542">
https://doi.org/10.5962/bhl.title.542
</ext-link>Examples
Linnæus C. 1758. Systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis Laurentii Salvii, Holmiae. Vol. Tomus I, Editio decima, reformata Edition: i-ii, 1-824. https://doi.org/10.5962/bhl.title.542
Recommendation
Each bibliographic citation must refer to the full bibliographic reference included in the List of references at the end of the article.
Each bibliographic reference should be complemented with a DOI. If no DOI exists, a DOI should be minted and added following rules and policies for retrospective DOI assignment at CrossRef and DataCite. When minting DOIs for legacy content, it's strongly recommended to carefully check and ensure that the article has not been assigned a DOI before, to avoid duplication.
All DOIs minted by a publisher, or anyone else, should be registered at the corresponding registration agency, CrossRef or DataCite, or other in case of legacy (BLR for instance).
Bibliographic citations in the text (e.g. Linnaeus 1758) should be cross-referenced to their bibliographic reference, mandatorily including DOIs for recently published articles, and hopefully for historical ones, in the backend article XML. This would allow easy harvesting and tracking of citations and discovery of the original literature source behind the reference.
Supplementary material
Definition
Supplementary material is used to add detail, background, or context to an article by providing backend data and information which are not formally part of the manuscript text, for example, multimedia objects such as audio clips and applets, raw data in a spreadsheet, additional XML-tagged sections, tables, or figures, or a source code of a software application in a repository*
What are identifiers for supplementary materials
CrossRef component DOIs, or DataCite DOIs, minted and submitted for registration by the publisher to CrossRef or DataCite, respectively.
How to discover supplementary materials’ identifiers
DOIs can be located using the search form provided by CrossRef or DataCite for their minted DOIs, or Refindit, which includes all the providers of DOIs.
How to mint identifiers for supplementary materials
CrossRef component DOIs are minted by the publishers, or on behalf of publishers and linked to the parent child DOI of the article. DataCite DOIs are minted by the DataCite partnering organisations and can be taken from there by the data aggregators and publishers.
How to annotate and cite supplementary materials
Cite: The supplementary material’s component DOI is linked to the parent DOI of the article (see example below) but can also be hyperlinked and made accessible independently from the article DOIs. It can be cited using the community accepted citing convention for data which should include its DOI, e.g., https://doi.org/10.3897/bdj.5.e14650.suppl1.
Annotate in JATS:
<ext-link
ext-link-type="doi"
xlink:href="https://10.3897/bdj.5.e14650.suppl1">
10.3897/bdj.5.e14650.suppl1
</ext-link>Examples
Van Achterberg K, Taeger A, Blank S, Zwakhals K, Viitasaari M, Yu D, de Jong Y (2017) Supplementary material 1 from: van Achterberg K, Taeger A, Blank S, Zwakhals K, Viitasaari M, Yu D, de Jong Y (2017) Fauna Europaea: Hymenoptera – Symphyta & Ichneumonoidea. Biodiversity Data Journal 5: e14650. https://doi.org/10.3897/BDJ.5.e14650. https://doi.org/10.3897/bdj.5.e14650.suppl1
In JATS XML, this example is annotated as:
<supplementary-material id="S3638143" orientation="portrait" position="float">
<object-id object-id-type="arpha">52DE64E0-3C7A-52B0-B88B-804A668FA4D4</object-id>
<object-id object-id-type="doi">10.3897/BDJ.5.e14650.suppl1</object-id>
<object-id object-id-type="zenodo_dep_id">904725</object-id>
<label>Supplementary material 1</label>
<caption>
<p>
<tp:taxon-name>
<tp:taxon-name-part taxon-name-part-type="superfamily">
Ichneumonoidea
</tp:taxon-name-part>
</tp:taxon-name>
— Fauna Europaea mapping
</p>
</caption>
<p>Data type: xlsx</p>
<p>Brief description: Cross-validation of Fauna Europaea (version 2.6.2) and Taxapad
(version 2016). Discrepancies are annotated. For details on data ownership and correct
citation please check the Fauna Europaea and Taxapad Websites.
</p>
<p>File: oo_145108.xlsx</p>
<media xlink:href="bdj-05-e14650-s001.xlsx"
mimetype="Microsoft Excel Document" mime-subtype="xlsx" position="float"
orientation="portrait">
<uri content-type="original_file">https://binary.pensoft.net/file/145108</uri>
</media>
<attrib specific-use="authors">Kees van Achterberg, Dicky Sick Ki Yu and Yde de Jong</attrib>
</supplementary-material>Recommendation
All metadata of supplementary material should be available in a standard, machine-readable format in the article backend XML and in human-readable citation formats suggested by the publisher at the article webpage.
Use CrossRef component DOI to identify each supplementary material files related to an article. The component DOI has the important feature to link the supplementary files DOI to its parent article DOI. If no CrossRef DOI are available, use DOIs from DataCite.
Conclusions
Over the last decades, the communities and the relevant scientific networks have realised the benefits of using PIDs, and they are developing initiatives to expand their use in other components of the publications. Adoption of identifiers in the scientific literature is a stepwise process and it may happen in two different ways: (1) data and sub-article level content is liberated from publications and identifiers are assigned to it retrospectively, or (2) they are prospectively published where the datum identifiers are provided upfront, at the moment of publication, and are available from the articles themselves, mostly from their published backend XML versions. These identifiers should be aligned through standards and community norms, for which the present paper provides guidance, and then re-used by data aggregators, for example, GBIF, Biodiversity Literature Repository, TreatmentBank, ARPHA-XML, OpenBiodiv, SIBiLS and others. This is leading to a unique and large corpus of FAIR literature data which can be linked to their original data sources and/or re-used for generation of new knowledge.
Though the development of semantically enhanced publications in taxonomy is much advanced, especially when compared to some much better funded science branches, such as molecular biology or ecology, its wider adoption in the publishing world is still to come due to a combination of technical and sociological issues. The composition and granularity of annotated sub-article elements discussed here are the first steps to providing machine access to the rich data in publications that would facilitate further exploration.
Future steps could include more structured geographic data or species traits (
To summarise, our recommendations presented in detail in the article text and in a concise form in Suppl. material
-
Persistent identifiers should be used as widely as possible for article metadata and sub-article structural elements and data.
-
Global unique persistent resolvable identifiers (GUPRI) provided by established international organisations such as CrossRef, DataCite, ORCID, INSDC, GBIF, etc., should always be preferred over locally assigned identifiers (UUIDs).
-
Persistent identifiers should be incorporated in the backend article XML to the maximum extent possible. The best practice in doing this is GUPRIs to be assigned as two different properties to an element: 1) as a “plain” UUID, and 2) as a resolvable UUID, that is, including its HTTP prefix. However more than one property for a PID per element should be allowed by the XML schema used. In case only one property per PID is allowed, then it is preferable to use the GUPRI or some other kind of resolvable PID instead of a UUID alone.
-
Assigning a persistent identifier to a named entity adds a semantic layer to it, however semantics and identifications do not always overlap. There are many cases when there is no need to add a PID to a sub-article element, however this element should still be appropriately tagged in the backend article XML. This is because much of the structure of the text, for example that of taxonomic treatments, implies the semantics needed for machine actionability and processing of the content.
-
In the world of biodiversity, assigning PIDs to data or other information entities should be aligned whenever possible with the current Darwin Core vocabulary and terms and other standards accepted by the biodiversity community in the Biodiversity Information Standards (TDWG) process. In Darwin Core coded data, PIDs should be placed consistently in the appropriate data fields intended for their use and management.
The data published in scholarly literature is normally considered as high-quality data, at least because it passes a review process and editorial evaluation and also because scientists associate their names and authority with the quality of data and content they publish. Hence, the data published in the literature is of special value to researchers, and to science as a whole, therefore it needs much more attention and exploration in the Internet-era compared to when text annotation and extraction tools did not exist.
The next important step is to convince more publishers, research infrastructures and biodiversity researchers to follow some or all of these guidelines, to achieve a complete integration of the published literature in the research lifecycle, not only in its usual human-readable form, but even more importantly, as data liberated from the narrative and re-imported back into the data lifecycle. For that goal, it is important that all actors in the research and publishing domains contribute to this process. For example:
-
Publishers need to implement semantic technologies in the publishing process that will facilitate text conversion to structured data. By doing this, they will benefit from higher visibility, citability and re-use of the content they publish.
-
Biodiversity research infrastructures participating in the BiCIKL and, more generally, in the alliance for biodiversity knowledge process, should commit to reusing the data extracted from literature by providing linkages between their source and related published data. A good example for that would be if INSDC automatically linked any mention of a particular sequence in the literature, or GBIF linked any mention of a specimen in the literature to its respective specimen record on their infrastructure. By doing this, research infrastructures will enrich their content, link it to other valuable sources of information and benefit their users by providing additional incentives to publish structured data.
-
Researchers, when writing their manuscripts, should use PIDs for data or sub-article elements whenever possible, and insist that publishers keep these PIDs in the published articles. By doing this, they will benefit from far richer sources of data from the published literature provided to them in a structured form and with less effort, gain higher visibility of their work and hopefully, increased opportunities for collaboration with other researchers.
-
All involved stakeholders, by implementing all or some of the recommendations in their daily work, should anticipate others to follow by demonstrating the impact of the identifiers in place, for example, by providing cases of generation of new knowledge based on data reuse, or by using PIDs to build alternative metrics to measure scientific output.
Acknowledgements
Financial support has been provided by the European Union's Horizon 2020 Biodiversity Integrated Knowledge Library (BiCIKL) project, under grant agreement No 101007492. This paper also benefitted from various discussion through the CETAF E-Publishing working group. The authors are grateful for the opportunity of such network and project to make room for fruitful and intensive discussions. Finally, the following colleagues provided valuable input that led to improve the manuscript: Anton Güntsch, Alex Ioannidis, Connie Rinaldo, and Clément Oury. The authors are grateful for their input and thank them for it.
References
- The Global Taxonomy Initiative in Support of the Post-2020 Global Biodiversity Framework.CBD Technical Seriies No. 96https://doi.org/10.5281/zenodo.5728812
- ‘openDS’ – Progress on the New Standard for Digital Specimens.Biodiversity Information Science and Standards4https://doi.org/10.3897/biss.4.59338
- Taxonomic information exchange and copyright: the Plazi approach.BMC Research Notes2(1). https://doi.org/10.1186/1756-0500-2-53
- Abb. 3 in Herausfinden, was wir schon wissen.Zenodohttps://doi.org/10.5281/zenodo.6998427
- Historia Animalium (English Translation). URL: http://classics.mit.edu/Aristotle/history_anim.5.v.html
- The international nucleotide sequence database collaboration.Nucleic Acids Research49(D1):D121‑D124. https://doi.org/10.1093/nar/gkaa967
- Proposals to provide for registration of new names and nomenclatural acts.Taxon65(3):656‑658. https://doi.org/10.12705/653.37
- Report of the Special Committee on registration of algal and plant names (including fossils).Taxon65(3):670‑672. https://doi.org/10.12705/653.43
- Joint statement on best practices for the citation of authorities of scientific names in taxonomy by CETAF, SPNHC and BHL.Research Ideas and Outcomes8https://doi.org/10.3897/rio.8.e94338
- Consortium of European Taxonomic Facilities (CETAF) best practices in electronic publishing in taxonomy.European Journal of Taxonomy475https://doi.org/10.5852/ejt.2018.475
- New species of the genus Chimarra Stephens from Africa (Trichoptera, Philopotamidae) and characterization of the African groups and subgroups of the genus.ZooKeys1111:43‑198. https://doi.org/10.3897/zookeys.1111.77586
- Taxonomic revision of the mydas-fly genera Eremohaplomydas Bequaert, 1959, Haplomydas Bezzi, 1924, and Lachnocorynus Hesse, 1969 (Insecta, Diptera, Mydidae).African Invertebrates63(1):19‑75. https://doi.org/10.3897/afrinvertebr.63.76309
- Three new species of Xiphocentron Brauer, 1870 (Trichoptera, Xiphocentronidae) from Mexico.ZooKeys(Special Issue in Honor of Ralph W. Holzenthal for a Lifelong Contribution to Trichoptera Systematics). https://doi.org/10.3897/zookeys.1111.73371
- Proceedings.Journal Article Tag Suite Conference (JATS-Con),Bethesda (MD), USA. https://doi.org/10.5281/zenodo.3484285
- EJT editorial standard for the semantic enhancement of specimen data in taxonomy literature.European Journal of Taxonomy586:1‑22. https://doi.org/10.5852/ejt.2019.586
- Revision of Chondrocyclus s.l. (Mollusca: Cyclophoridae), with description of a new genus and twelve new species.European Journal of Taxonomy569https://doi.org/10.5852/ejt.2019.569
- Time to change how we describe biodiversity.Trends in Ecology & Evolution27(2):78‑84. https://doi.org/10.1016/j.tree.2011.11.007
- Making the world a PIDder place: It's up to all of us!Zenodo.DataCite Member Meeting 2021, virtual. https://doi.org/10.5281/zenodo.5532631
- A best practice guide for semantic enhancement and improvement of semantic interoperability.DiSSCo Prepare deliverable 5.4 report. https://doi.org/10.34960/ajxs-zr25
- Infrastructure and Population of the OpenBiodiv Biodiversity Knowledge Graph.Biodiversity Data Journal9https://doi.org/10.3897/bdj.9.e67671
- Solutions for a sustainable EOSC. A FAIR Lady (olim Iron Lady) report from the EOSC Sustainability Working Group. https://doi.org/10.2777/870770
- A Persistent Identifier (PID) policy for the European Open Science Cloud (EOSC).Report from the European Open Science Cloud FAIR and Architecture Working Groups. https://doi.org/10.2777/926037
- Digital accessible knowledge: Mobilizing legacy data and the future of taxonomic publishing.Bulletin of the Society of Systematic Biologists1(1):1‑12. https://doi.org/10.18061/bssb.v1i1.8296
- Principles for creating a single authoritative list of the world’s species.PLoS Biology18(7). https://doi.org/10.1371/journal.pbio.3000736
- Synospecies, an application to reflect changes in taxonomic names based on a triple store based on taxonomic data liberated from publication.Biodiversity Information Science and Standards5https://doi.org/10.3897/biss.5.75641
- People are essential to linking biodiversity data.Database2020https://doi.org/10.1093/database/baaa072
- The disambiguation of people names in biological collections.Biodiversity Data Journal10https://doi.org/10.3897/bdj.10.e86089
- Connecting molecular sequences to their voucher specimens.BioHackrXivhttps://doi.org/10.37044/osf.io/93qf4
- Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects.Database2017https://doi.org/10.1093/database/bax003
- A botanical demonstration of the potential of linking data using unique identifiers for people.PLoS One16(12). https://doi.org/10.1371/journal.pone.0261130
- The Trouble with Triplets in Biodiversity Informatics: A Data-Driven Case against Current Identifier Practices.PLoS ONE9(12). https://doi.org/10.1371/journal.pone.0114069
- Community next steps for making globally unique identifiers work for biocollections data.ZooKeys494:133‑154. https://doi.org/10.3897/zookeys.494.9352
- ‘openDS’ – A New Standard for Digital Specimens and Other Natural Science Digital Object Types.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37033
- A choice of persistent identifier schemes for the Distributed System of Scientific Collections (DiSSCo).Research Ideas and Outcomes7https://doi.org/10.3897/rio.7.e67379
- Digital Extended Specimens: Enabling an Extensible Network of Biodiversity Data Records as Integrated Digital Objects on the Internet.BioScience72(10):978‑987. https://doi.org/10.1093/biosci/biac060
- Towards a global list of accepted species VI: The Catalogue of Life checklist.Organisms Diversity & Evolution21(4):677‑690. https://doi.org/10.1007/s13127-021-00516-w
- Stable citations for herbarium specimens on the internet: an illustration from a taxonomic revision of Duboscia (Malvaceae).Phytotaxa73(1). https://doi.org/10.11646/phytotaxa.73.1.4
- Ardisia whitmorei (Primulaceae-Myrsinoideae), a new species from north east of Peninsular Malaysia.PhytoKeys204:35‑41. https://doi.org/10.3897/phytokeys.204.86647
- Registration of Algal Novelties in Phycobank: Serving the scientific community and filling gaps in the global names backbone.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37285
- FAIR Data and Services in Biodiversity Science and Geoscience.Data Intelligence2:122‑130. https://doi.org/10.1162/dint_a_00034
- Species plantarum : exhibentes plantas rite cognitas ad genera relatas, cum diferentiis specificis, nominibus trivialibus, synonymis selectis, locis natalibus, secundum systema sexuale digestas.Holmiæ, impensis Laurentii Salvii. https://doi.org/10.5962/bhl.title.37656
- Systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis Laurentii Salvii, Holmiae.Editio decima, reformata Edition: i-iiVol. Tomus I:1‑824. https://doi.org/10.5962/bhl.title.542
- Persistent Identifiers at the Natural History Museum.British Libraryhttps://doi.org/10.22020/k99s-we61
- Rebuilding community ecology from functional traits.Trends in Ecology & Evolution21:178‑185. https://doi.org/10.1016/j.tree.2006.02.002
- Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data.PLoS Biol15(6). https://doi.org/10.1371/journal.pbio.2001414
- The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications.Nucleic Acids Research47:259‑264. https://doi.org/10.1093/nar/gky1022
- Surfacing the deep data of taxonomy. In: Michel E (Ed.) Anchoring Biodiversity Information: From Sherborn to the 21st century and beyond.ZooKeys550:247‑260. https://doi.org/10.3897/zookeys.550.9293
- Ozymandias: A biodiversity knowledge graph.Zenodohttps://doi.org/10.5281/zenodo.2634326
- TraitBank: Practical semantics for organism attribute data.Semantic Web7:577‑588. https://doi.org/10.3233/SW-150190
- Records of Wenchengia (Lamiaceae) from Vietnam.Biodiversity Data Journal4:9596. https://doi.org/10.3897/BDJ.4.e9596
- Evolutionary relationships and population genetics of the Afrotropical leaf-nosed bats (Chiroptera, Hipposideridae).ZooKeys929:117‑161. https://doi.org/10.3897/zookeys.929.50240
- Scientific names of organisms: attribution, rights, and licensing.BMC Research Notes7(79). https://doi.org/10.1186/1756-0500-7-79
- Semantic tagging of and semantic enhancements to systematics papers: ZooKeys working examples.ZooKeys50:1‑16. https://doi.org/10.3897/zookeys.50.538
- XML schemas and mark-up practices of taxonomic literature.ZooKeys150:89‑116. https://doi.org/10.3897/zookeys.150.2213
- Implementation Of Taxpub, An Nlm Dtd Extension For Domain-Specific Markup In Taxonomy, From The Experience Of A Biodiversity Publisher.Zenodohttps://doi.org/10.5281/zenodo.804247
- A common registration-to-publication automated pipeline for nomenclatural acts for higher plants (International Plant Names Index, IPNI), fungi (Index Fungorum, MycoBank) and animals (ZooBank).ZooKeys550:233‑246. https://doi.org/10.3897/zookeys.550.9551
- OpenBiodiv: A Knowledge Graph for Literature-Extracted Linked Open Data in Biodiversity Science.Zenodo7(2):38. https://doi.org/10.5281/zenodo.4061905
- Biodiversity Community Integrated Knowledge Library (BiCIKL).Research Ideas and Outcomes8:81136. https://doi.org/10.3897/rio.8.e81136
- A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System.PLoS ONE8(8):66213. https://doi.org/10.1371/journal.pone.0066213
- International code of zoological nomenclature.Fourth edition. URL: https://www.iczn.org/the-code/the-international-code-of-zoological-nomenclature/
- Towards an ecological trait‐data standard.Methods in Ecology and Evolution10:2006‑2019. https://doi.org/10.1111/2041-210X.13288
- NCBI Taxonomy: a comprehensive update on curation, resources and tools.Database2020https://doi.org/10.1093/database/baaa062
- OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system.Journal of Biomedical Semantics9(5). https://doi.org/10.1186/s13326-017-0174-5
- Monomorium dryhimi sp. n., a new ant species (Hymenoptera, Formicidae) of the M. monomorium group from Saudi Arabia, with a key to the Arabian Monomorium monomorium-group.ZooKeys106:47‑54. https://doi.org/10.3897/zookeys.106.1390
- The NCBI BioCollections Database.Database2018https://doi.org/10.1093/database/bay006
- International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017.Regnum Vegetabile 159. Glashütten, Koeltz Botanical Books. https://doi.org/10.12705/Code.2018
- Liberating host-virus knowledge from biological dark data.The Lancet Planetary Healthhttps://doi.org/10.1016/S2542-5196(21)00196-0
- Let the concept of trait be functional!Oikos116:882‑892. https://doi.org/10.1111/j.0030-1299.2007.15559.x
- Aristotle’s scientific contributions to the classification, nomenclature and distribution of marine organisms.Mediterranean Marine Science18:468‑478. https://doi.org/10.12681/mms.13874
- Semantics in support of biodiversity knowledge discovery: an introduction to the biological collections ontology and related ontologies.PloS one9(3):89606. https://doi.org/10.1371/journal.pone.0089606
- Describing species: Practical taxonomic procedures for biologists.Zenodohttps://doi.org/10.5281/zenodo.7006541
- Two new species of Neopestalotiopsis from southern China.Biodiversity Data Journal9https://doi.org/10.3897/bdj.9.e70446
- A new species of giant Eunice (Eunicidae, Polychaeta, Annelida) from the east coast of Australia.ZooKeys1118:97‑109. https://doi.org/10.3897/zookeys.1118.86448
- Two new Halamphora (Bacillariophyta) species from the marine coasts off Livingston Island, Antarctica.PhytoKeys195:161‑174. https://doi.org/10.3897/phytokeys.195.81632
Supplementary material
As the most recent version of TaxPub (V1rc2) extends (https://github.com/plazi/TaxPub/) the JATS Journal Publishing Tag Set Version 1.1 (https://jats.nlm.nih.gov/publishing/1.1/) that version is referred to in the table. In the forthcoming version of TaxPub.
CETAF Stable Identifiers https://cetaf.org/resources/bestpractices/
Thiers, B. M. (updated continuously). Index Herbariorum. http://sweetgum.nybg.org/science/ih/
Besides people, Wikidata covers many institutions and collections too, and identifiers for them can be minted and used in essentially the same way as described for persons above.
Sample queries for funding acknowledgments and general acknowledgments: https://scholia.toolforge.org/sponsor/Q304878 and https://w.wiki/5XnJ