|
Research Ideas and Outcomes :
Project Report
|
|
Corresponding author: Sofie Meeus (sofie.meeus@plantentuinmeise.be)
Received: 10 Oct 2022 | Published: 14 Oct 2022
This is an open access article distributed under the terms of the CC0 Public Domain Dedication.
Citation:
Meeus S, Addink W, Agosti D, Arvanitidis C, Balech B, Dillen M, Dimitrova M, González-Aranda JM, Holetschek J, Islam S, Jeppesen TS, Mietchen D, Nicolson N, Penev L, Robertson T, Ruch P, Trekels M, Groom Q (2022) Recommendations for interoperability among infrastructures. Research Ideas and Outcomes 8: e96180. https://doi.org/10.3897/rio.8.e96180
|
|
Abstract
The BiCIKL project is born from a vision that biodiversity data are most useful if they are presented as a network of data that can be integrated and viewed from different starting points. BiCIKL’s goal is to realise that vision by linking biodiversity data infrastructures, particularly for literature, molecular sequences, specimens, nomenclature and analytics. To make those links we need to better understand the existing infrastructures, their limitations, the nature of the data they hold, the services they provide and particularly how they can interoperate. In light of those aims, in the autumn of 2021, 74 people from the biodiversity data community engaged in a total of twelve hackathon topics with the aim to assess the current state of interoperability between infrastructures holding biodiversity data. These topics examined interoperability from several angles. Some were research subjects that required interoperability to get results, some examined modalities of access and the use and implementation of standards, while others tested technologies and workflows to improve linkage of different data types.
These topics and the issues in regard to interoperability uncovered by the hackathon participants inspired the formulation of the following recommendations for infrastructures related to (1) the use of data brokers, (2) building communities and trust, (3) cloud computing as a collaborative tool, (4) standards and (5) multiple modalities of access:
-
If direct linking cannot be supported between infrastructures, explore using data brokers to store links
-
Cooperate with open linkage brokers to provide a simple way to allow two-way links between infrastructures, without having to co-organize between many different organisations
-
Facilitate and encourage the external reporting of issues related to their infrastructure and its interoperability.
-
Facilitate and encourage requests for new features related to their infrastructure and its interoperability.
-
Provide development roadmaps openly
-
Provide a mechanism for anyone to ask for help
-
Discuss issues in an open forum
-
Provide cloud-based environments to allow external participants to contribute and test changes to features
-
Consider the opportunities that cloud computing brings as a means to enable shared management of the infrastructure.
-
Promote the sharing of knowledge around big data technologies amongst partners, using cloud computing as a training environment
-
Invest in standards compliance and work with standards organisations to develop new, and extend existing standards
-
Report on and review standards compliance within an infrastructure with metrics that give credit for work on standard compliance and development
-
Provide as many different modalities of access as possible
-
Avoid requiring personal contacts to download data
-
Provide a full description of an API and the data it serves
Finally, the hackathons were an ideal meeting opportunity to build, diversify and extend the BiCIKL community further, and to ensure the alignment of the community with a common vision on how best to link data from specimens, samples, sequences, taxonomic names and taxonomic literature.
Keywords
hackathon, biodiversity informatics, API, Wikidata, cloud computing, data standards, FAIR data, linking
Preface
Providing services to science through data infrastructures is a complex and challenging job that requires juggling often conflicting needs of users, future developments, routine maintenance and software lifecycles. With all these pressures it is perhaps difficult to step back and evaluate where investment is needed and what the future opportunities are. This is one of the reasons that a hackathon was chosen as a mechanism to examine the interoperability of infrastructures (Suppl. materials
Introduction
The overarching goal of BiCIKL is to create a community of infrastructures concerned with data on biodiversity through liberating data from scholarly publications and bi-directional linking of literature, taxonomic, DNA sequence and occurrence data (
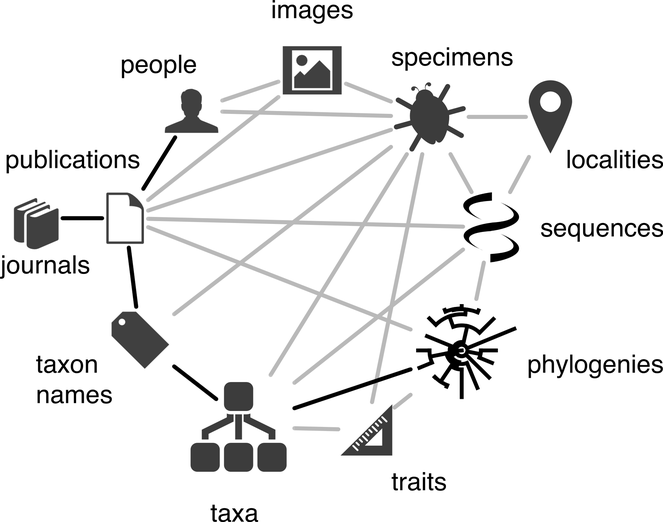
At a technical level BiCIKL intends to achieve its goals through the provision of data, tools and services to the community. It will cover the whole research life cycle and will contribute new methods and workflows to harvest, liberate, link, reuse data from specimens, samples, sequences, taxonomic names and taxonomic literature (Fig.
A diagram of the biodiversity knowledge graph taken from
Undoubtedly, the pandemic has presented a challenge to collaborative working, and particularly a hackathon that pre-pandemic was defined by the radial collocation of its participants (
A hackathon is an event of limited duration where teams tackle technical problems together, test ideas, create solutions, learn new skills, socialise and discuss (
We also participated in the Biohackathon 2021. BioHackathons have been organised for almost twenty years to take advantage of the hackathon format in the life sciences (
Everyone from the BiCIKL community was encouraged to submit topics for pilot projects to test interoperability between the infrastructures. The topics were retrospectively grouped into three themes
- research-based questions,
- evaluating the infrastructures’ modalities of access, and the use and implementation of standards, and
- testing technologies and workflows to improve linkage of different data types.
Below, we outline these topics and use them to support five high-level recommendations for infrastructures to improve their interoperability.
Recommendations to the infrastructures
1. Use of data brokers
In principle data infrastructures can be linked directly together. Stable identifiers of digital entities on one infrastructure can be maintained on another to link infrastructures in one direction, or there can be reciprocal links to traverse infrastructures in either direction. Indeed, such linkage is implied by the knowledge graph depicted in Fig.
An alternative to linking infrastructures is for a third party infrastructure to act as a broker between infrastructures. Wikidata is a collaboratively edited multilingual database hosted by the Wikimedia foundation (
There are several advantages of data brokerage through Wikidata in addition to direct linking. The broker infrastructure has an incentive to maintain the links, because that is a primary function of that infrastructure. Wikidata is open to editing from anyone, which both allows users to contribute and correct links, but it also means the people that need the links are incentivized to provide them. At first sight it seems that a data broker adds an additional point of failure and additional search and processing requirements. However, a data broker can link many infrastructures together simultaneously meaning that one additional broker system can join a whole family of infrastructures together. The main requirement is for infrastructures to keep their key identifiers stable, but there is clearly an incentive to maintain stable identifiers if those identifiers help link the infrastructure in both directions to a host of other data.
RECOMMENDATIONS
- If direct linking cannot be supported between infrastructures, explore using data brokers to store links.
- Cooperate with open linkage brokers to provide a simple way to allow two-way links between infrastructures, without having to co-organize between many different organisations.
2. Building communities/trust
BiCIKL is a project about building a community and trust between infrastructures is an essential aspect of interoperability that goes beyond the purely technical issues. If infrastructures are going to invest resources to interoperate with each other they need to know that the other infrastructures will use the systems and standards that are put in place; that they will be consulted on the design and implementation and that there will be sufficient stability that the interoperability will last, such as ensuring backwards compatibility.
The community, however, extends beyond the infrastructures to the users, whether they are data providers or downstream consumers of the infrastructure’s services. The user community will not only make use of the linked infrastructures but will also contribute to it, for example, by enriching data brokers and providing user feedback to infrastructures. The infrastructures should facilitate the reporting of issues, including those issues related to incompatibilities between infrastructures. Good examples of issue tracking are in place, but need to be visible to the users and issues should be responded to promptly and constructively. GitHub is often used as an issue tracker and the ability to discuss, prioritise and label issues are important to building trust. Nevertheless, not everyone is comfortable using GitHub so if the infrastructure has a large number of non-informatics users then other forms of feedback and issue tracking might be necessary. Some infrastructures also provide a user forum where users can ask questions and debate issues. Such fora can be invaluable for providing support, self help and can be a place new features can be discussed. There are also many external fora where infrastructure services are discussed and it makes sense for these to be monitored by the infrastructures as a means to understand their community.
An important aspect to community building is that potential community members recognize other people in the community with common skills, needs and experience. So while preparing the hackathon we paid particular attention to the demographic and diversity of skills of the participants. For example, hackathons can tend to be biassed towards male participation (
For example, topic 5 (Suppl. material
Having an Open Source code-base might be another way that users could resolve their own issues within the community. All of the above build trust between infrastructures and between infrastructures and users. This builds engagement, avoids infrastructure being reinvented, supports both technical and social innovation, and is inclusive.
Technology can also be used to underpin trust in infrastructures (
RECOMMENDATIONS
- Facilitate and encourage the external reporting of issues related to their infrastructure and its interoperability.
- Facilitate and encourage requests for new features related to their infrastructure and its interoperability.
- Provide development roadmaps openly.
3. Cloud computing as a collaborative tool
Cloud Computing technology provides the means for system developers to purchase computation and storage resources for a period of time without the need to acquire or manage physical hardware. This can bring real benefit under some scenarios, such as the need for high computation capacity for short periods of time, to scale a system up with growing demand or performing tests using different hardware configurations. The growing maturity of cloud computing services available, such as from Amazon and Microsoft now provide easy to use tools that enable a small team to quickly manage complex environments. Having access to this capability, along with recipes and tutorials for managing aspects like security and backup is an attractive proposition for any team.
An important aspect of cloud computing that is attractive to the BiCIKL project is the ability to collaborate. The infrastructures connected to BiCIKL are typically operated on an institutional network with limited possibility for external collaborators to get involved. Even though the software is often developed in an open source manner, it can be near impossible for an external person to reproduce the environment and contribute significantly. During the BiCIKL hackathon a portion of the GBIF infrastructure was recreated on the Microsoft Azure cloud for topic 3 (Suppl. material
Beyond collaboration, cloud infrastructures also commonly offer various services built on massive-scale Machine Learning implementations. This includes powerful enrichment services such as georeferencing, computer vision, translating and data clustering. Infrastructures may make use of such state-of-the-art services to enrich the data they serve and make links to other infrastructures, benefitting from a scaling effectiveness they could not meet on their own. An example is handwritten text recognition for sparse and high variance text lines, such as occur regularly on scanned labels (topic 12, Suppl. material
Importantly, it should be noted that cloud computing comes at a financial cost, which may be offset through grants offering free credit. The costs of operating the Azure cloud for this hackathon was funded through a grant from the Microsoft Planetary Computer programme. Computer Vision-based linking approaches were piloted on voucher credit, but could be quite costly if implemented on a larger scale.
RECOMMENDATIONS
- Provide cloud-based environments to allow external participants to contribute and test changes to features.
- Consider the opportunities that cloud computing brings as a means to enable shared management of the infrastructure.
- Promote the sharing of knowledge around big data technologies amongst partners, using cloud computing as a training environment.
4. Standards
It is a fairly obvious statement that adoption and continued compliance with community standards is a positive step towards interoperability (cf. FAIR principles; Wilkinson et al. 2016). Standards include the use of common terms, controlled vocabularies and also data models. Standards are not, and should not be, static instruments of interoperability. They provide meaning and structure to data, but they also influence the types and resolution of the data collected. Therefore, they are not independent of the intended uses of data, which leads to some of the disparities between competing standards and incomparable implementations of common standards. In cases where a small community is trying to connect with a larger one, adoption of the larger community's standards is a good first step. For example, the use of IIIF in topic 9 (Suppl. material
As a case where standards are failing, topic 1 (Suppl. material
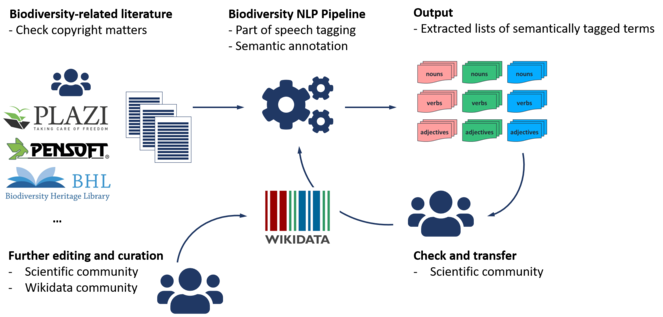
In the case of taxa and taxon names topic 12 (Suppl. material
Looking forward to the future of biodiversity standards, the FAIR Digital Object topic 6 (Suppl. material
Standards need to be developed by a broad community to be useful to that whole community. But standards development and compliance need investment by infrastructures. Although widespread standards compliance across infrastructures would significantly enhance interoperability there are limitations to how far standard compliance can go. The primary objectives of the infrastructure come first and standards compliance has to compete for resources with other priorities. Nevertheless, there is a risk that infrastructure managers fail to see the potential for new users and uses of the infrastructure, because without standards compliance these potential users and uses are blocked and are therefore invisible.
RECOMMENDATIONS
- Invest in standards compliance and work with standards organisations to develop new, and extend existing standards.
- Report on and review standards compliance within an infrastructure with metrics that give credit for work on standard compliance and development.
5. Modalities of access
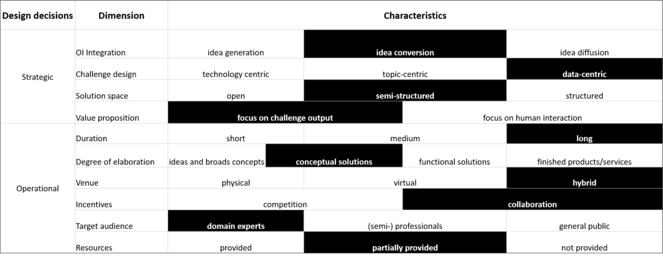
The ways that researchers access data can have a large influence on what research is conducted and how easy it is for researchers to do what they want. BiCIKL infrastructures aim to provide Open Data to be used however the users want. They want to support innovative uses and novel applications, but also more prosaic uses for the data. The aim is to do more and better science in a timely manner. The modes by which data are accessed is an important consideration in reducing the barriers and friction to use of these data. They are also critical to what uses can be made of the data. We recommend that infrastructures provide as many different modalities of access as possible. Only by doing this will they give access to the data without limiting the uses that researchers can make of the data. We have distinguished four basic levels of access, all of which have use to the community. These are:
- browsing the data via a web portal,
- programmatic access via an API,
- downloading data to be used locally and
- personal requests for unique sets of data.
In the hackathon topics all of these modes were used (Fig.
Portal Access
Web portal access to the data allows users to evaluate what data is available in an infrastructure, in what format and what the quality and structure is like. They also support simple information requests. They are usually the first point of contact a researcher has with an infrastructure and are therefore critical to supporting a longer relationship with that researcher. If web portal access is slow, confusing or incomplete it is likely that the potential user will either go elsewhere or create their own resources.
Application Programming Interfaces (APIs)
Web APIs provide simple programmatic access to data. They can be built into workflows and made completely automatic and repeatable, keeping the output up-to-date with the latest data in the infrastructure. Tools can be built upon them to connect and retrieve information from multiple infrastructures at once (Suppl. material
Personal requested data
A feature of several hackathon topics was the use of data provided from an infrastructure through personal contact with one of the administrators. This was to circumvent the limitations of the modalities of access provided, such as where a public API or download facility is not provided, or those facilities do not provide access to all the data or the data are in an unsuitable format. Personally requested data are sometimes necessary, but they are also an indication that there is an unresolved demand for access from users. It is very useful to researchers if infrastructures can support them with bespoke requests, however they are also problematic from several stand points. Such requests may only be possible due to personal contacts of the researcher with those in the infrastructure. This does not allow a level playing field for research. It is an inefficient way to provide data and it does not support reproducibility and citation, because it is more difficult to track provenance.
Downloads
Data science often requires large amounts of data to be analysed and the only way to process these data efficiently is to create a local copy. Infrastructures should provide download access to all or part of the data so that it can be easily retrieved by researchers. This could be provided in several ways. GBIF provides an asynchronous download system for queries and direct downloads of individual datasets. In the absence of a dedicated download system users may try to achieve the same result through an API, but this is highly inefficient for the user and infrastructure.
RECOMMENDATIONS
- Provide as many different modalities of access as possible.
- Avoid requiring personal contacts to download data.
- Provide a full description of an API and the data it serves.
Acknowledgements
The authors thank all the participants and their respective organisations for their contributions in the hackathons (see participant list in Appendix). In particular, we would like to thank the five invitees to the BiCIKL hackathon at Meise Botanic Garden: Christine Driller, Marina Golivets, Rukaya Johaadien, Sarah Vincent and Sabine von Mering for their participation and valuable contributions.
Funding program
The BiCIKL hackathon and participation in the Biohackathon Europe was funded by the BiCIKL project (European Union’s Horizon 2020 Research and Innovation Action under grant agreement No 101007492), DiSSCo Flanders (Research Foundation – Flanders research infrastructure under grant number FWO I001721N) and the Elixir BioHackathon Europe. Microsoft provided Azure cloud computing credit through the Planetary Computer programme to support the collaboration in topic 3.
Mariya Dimitrova was supported with funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 764840.
Hosting institution
Meise Botanic Garden, Belgium
References
- What Do We Know About Hackathon Outcomes and How to Support Them? – A Systematic Literature Review. In:Lecture Notes in Computer Science,12324.International Conference on Collaboration Technologies and Social Computing.SpringerCollaboration Technologies and Social Computing,50–64pp. [ISBN978-3-030-58157-2]. https://doi.org/10.1007/978-3-030-58157-2_4
-
Digital innovation: The hackathon phenomenon.URL: http://qmro.qmul.ac.uk/xmlui/handle/123456789/11418
- Globally distributed object identification for biological knowledgebases.Briefings in bioinformatics5(1):59‑70. https://doi.org/10.1093/bib/5.1.59
- FAIR Digital Objects for Science: From Data Pieces to Actionable Knowledge Units.Publications8(2):21. https://doi.org/10.3390/publications8020021
- Ten simple rules to run a successful BioHackathon.PLOS Computational Biology16(5):1007808. https://doi.org/10.1371/journal.pcbi.1007808
- People are essential to linking biodiversity data.Database2020https://doi.org/10.1093/database/baaa072
- Connecting molecular sequences to their voucher specimens.BioHackrXivhttps://doi.org/10.37044/osf.io/93qf4
- Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects.Database2017:bax003. https://doi.org/10.1093/database/bax003
- The Trouble with Triplets in Biodiversity Informatics: A Data-Driven Case against Current Identifier Practices.PLoS ONE9(12). https://doi.org/10.1371/journal.pone.0114069
- What the Hack? – Towards a Taxonomy of Hackathons. In:Lecture Notes in Computer Science,11675.Business Process Management. BPM 2019.Springer,354–369pp. [ISBN978-3-030-26619-6]. https://doi.org/10.1007/978-3-030-26619-6_23
- Use of globally unique identifiers (GUIDs) to link herbarium specimen records to physical specimens.Applications in Plant Sciences6(2):1027. https://doi.org/10.1002/aps3.1027
- Towards a biodiversity knowledge graph.Research Ideas and Outcomes2https://doi.org/10.3897/rio.2.e8767
- Towards Interlinked FAIR Biodiversity Knowledge: The BiCIKL perspective.Biodiversity Information Science and Standards5https://doi.org/10.3897/biss.5.74233
- Biodiversity Community Integrated Knowledge Library (BiCIKL).Research Ideas and Outcomes8https://doi.org/10.3897/rio.8.e81136
- Understanding Hackathons for Science: Collaboration, Affordances, and Outcomes. In:Lecture Notes in Computer Science,11420.International Conference on Information.SpringerInformation in Contemporary Society. iConference 2019,27–37pp. [ISBN978-3-030-15742-5]. https://doi.org/10.1007/978-3-030-15742-5_3
- StitchFest: Diversifying a College Hackathon to Broaden Participation and Perceptions in Computing.46th ACM Technical Symposium on Computer Science Education,Kansas City Missouri,March 4 - 7, 2015.Association for Computing Machinery,New York,114–119pp. [ISBN978-1-4503-2966-8]. https://doi.org/10.1145/2676723.2677310
- Wikidata.Information Technology and Libraries38(2):72‑81. https://doi.org/10.6017/ital.v38i2.10886
- Wikidata: a new platform for collaborative data collection. ,Lyon, France,April 16-20, 2012.Association for Computing Machinery,New York,1063–1064pp. [ISBN9781450312301]. https://doi.org/10.1145/2187980.2188242
- The FAIR Guiding Principles for scientific data management and stewardship.Scientific Data3(1). https://doi.org/10.1038/sdata.2016.18
Supplementary materials
Description of hackathon topic 1.
Description of hackathon topic 2.
Description of hackathon topic 3.
Description of hackathon topic 4.
Description of hackathon topic 5.
Description of hackathon topic 6.
Description of hackathon topic 7.
Description of hackathon topic 8.
Description of hackathon topic 9.
Description of hackathon topic 10.
Description of hackathon topic 11.
Description of hackathon topic 12.
Group picture of on-site participants of the BiCIKL hackathon at Meise Botanic Garden.
Group picture of online participants of the BiCIKL hackathon at Meise Botanic Garden.
List of participants involved in the BiCIKL hackathon and participants in the BioHackathon Europe that worked on two biodiversity data related projects.