|
Research Ideas and Outcomes :
Conference Abstract
|
|
Corresponding author: Allen J Flynn (ajflynn@umich.edu)
Received: 03 Sep 2022 | Published: 12 Oct 2022
© 2022 Allen Flynn, Marisa Conte, Peter Boisvert, Rachel Richesson, Zach Landis-Lewis, Charles Friedman
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Flynn AJ, Conte M, Boisvert P, Richesson R, Landis-Lewis Z, Friedman CP (2022) Linked Metadata for FAIR Digital Objects Carrying Computable Knowledge. Research Ideas and Outcomes 8: e94438. https://doi.org/10.3897/rio.8.e94438
|
|
Abstract
Introduction
To advance the goals of the Mobilizing Computable Biomedical Knowledge (MCBK) Movement, we are exploring the use of FAIR Digital Objects (FDOs) (
First, we are beginning to clarify the full range of metadata for FDOs that carry bit sequences expressing knowledge in machine readable or executable formats. We view knowledge through an empirical lens as the reliable, valid, and valued results of analytic or deliberative data analysis. Computability of knowledge refers to the degree to which knowledge is formally represented for use by computing machines.
Second, we are figuring out how to apply linked data principles to FDO metadata records (
In keeping with the FAIR Digital Object Framework (FDOF), we value linked metadata as a general method of bringing consistency to FDO metadata records, making it so that artificial agents can act on them in predictable ways. Five other benefits of linked metadata are that they are divisible, aggregable, extensible, queryable (using SPARQL), and support logical inferencing.
With a focus specifically on FDOs that carry computable knowledge artifacts at their core, here we present our recent metadata work completed between 2019 and mid-2022.
Metadata Scope for FDOs Carrying Computable Knowledge
This section summarizes previously published work to specify and scope FDO metadata. This work was completed by members of our team and the larger MCBK Movement. Through many dialogs over a period of more than a year, thirteen high-level categories of metadata for FDOs carrying computable knowledge were described (
|
1. Type |
8. Authorization and Rights Management |
|
2. Knowledge Domain* |
9. Preservation |
|
3. Purpose* |
10. Integrity |
|
4. Identification |
11. Provenance |
|
5. Location |
12. Evidential Basis* |
|
6. FDO-to-FDO Relation* |
13. Evidence from Use* |
|
7. Technical* |
|
For detailed explanations and examples of each metadata category above, see our full publication.
Next, we briefly discuss six categories marked with an asterisk (*) in Table
For Knowledge Domain metadata, a large and growing number of biomedical vocabularies or schema exist. For clinical terms, the Standardized Nomenclature of Medicine (SNOMED) includes more than 350K RDF classes and 200 properties. Many bioscience vocabularies spanning a wide range of terms from human biology also exist.
Purpose metadata are critical for FDOs that convey computable knowledge about the prevention, diagnosis, treatment, amelioration, and monitoring of disease. Interestingly, we have yet to find vocabularies for representing clinically-oriented FDO purposes as linked metadata.
We anticipate needing FDO-to-FDO Relation metadata. Going beyond citations that relate knowledge to its antecedents, FDOs containing computable biomedical knowledge may relate sequentially (diagnostic knowledge preceding treatment knowledge), dependently (stratification depends on measurement), or comparatively (multiple models estimate the same factor). More work is needed to formalize these relations.
For technical metadata about FDOs carrying computable knowledge, we emphasize existing vocabularies, including software ontologies like the function ontology. Moreover, for certain FDO operations, webservices are a way of leveraging the decentralized web. As Technical FDO metadata, we can describe FDO-backed webservices semantically by building on the work of the OpenAPI and AsyncAPI initiatives.
Finally, we need FDO metadata about two different kinds of evidence. First, there are Evidential Basis metadata that describe features and details about how computable knowledge contained FDOs was generated. Second, there are Evidence from Use metadata that describe the effects of applying the computable knowledge contained in FDOs to simulated or real cases.
Linked Metadata for actual FDOs Carrying Computable Knowledge
This section shares new work. Since 2016, we have built and tested several hundred compound Digital Objects (DOs) carrying executable biomedical knowledge in the form of pure functions (e.g., math functions for estimating a health risk) (
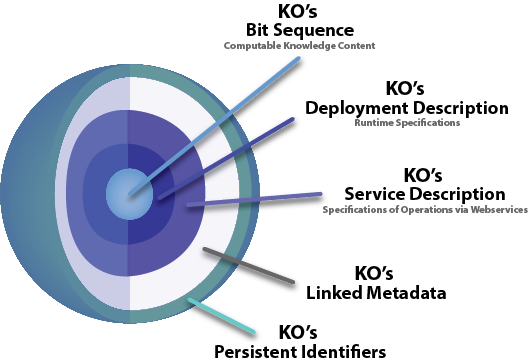
General Diagram of a Knowledge Object
This figure depicts the parts of a type of DOs called Knowledge Objects (KOs). The core of the KO is a bit sequence encoding some machine processable knowledge. This core is referred to as the KO’s payload. For all KOs, the payload can be deployed automatically on the web as a webservice by software tools that act on the KOs Deployment and Service Descriptions. The KO and its payload are described by metadata of different kinds. The KO has a persistent identifier (PID) that facilities gaining access to its components
In a move towards having a specific type of FDOs for carrying computable knowledge, we have started the process of developing linked metadata records for FDOs using a prototype metadata schema. An example of an early FDO linked data record appears in Example 1.
{ "@context": { "dcterms": "http://purl.org/dc/terms/", "koio": "http://kgrid.org/koio/", "fno" : "https://w3id.org/function/ontology/" }, "@id":"https://library.kgrid.org/#/object/99999%2Ffk4jh3tk9s%2Fv1.0%2Fv1.0", "@type": "koio:KnowledgeObject", "dcterms:title" : " Tammemagi, 6 year Lung Cancer Risk Prediction Model for Screening", "dcterms:identifier" : " ark:/99999/fk4jh3tk9s", "dcterms:hasVersion" :"v1.0", "dcterms:created":"2016-04-15", "dcterms:description" : "A 10-factor patient-level logistic regression model for estimating the risk of a future lung cancer diagnosis for a person", "dcterms:creator" : ["https://kgrid.org/ ","https://medicine.umich.edu/dept/learning-health-sciences"], "dcterms:source" : ["https://www.nejm.org/doi/pdf/10.1056/NEJMoa1211776"], "dcterms:publisher" : " https://medicine.umich.edu/dept/learning-health-sciences", "dcterms:rights" : "All rights reserved.", "dcterms:rightsHolder" : "Department of Learning Health Sciences, University of Michigan Medical School, 1111 E Catherine Street, Ann Arbor, MI, 48109", "dcterms:license":"NOT licensed for use outside the Department of Learning Health Sciences", "dcterms:valid" : "2016-04-15/2016-04-16", "dcterms:hasPart":["getSixyearprobability.js","deployment.yaml","service.yaml","metadata.jsonld"], "koio:hasPayload" : { "@id":"getSixyearprobability.js", "@type" : "fno:function", "dcterms:title" : " getSixyearprobability", "dcterms:language" : "Javascript", "fno:solves" : "Maps patient features to lung cancer risk scores", "fno:expects" : ["age", "ethnicity", "bmi","cigsPerDay","edLevel","hxLungCancer","hxLungCancerFam","hxNonLungCancerDz","yrsQuit","yrsSmoker"], "fno:returns" :["Lung Cancer Risk Score"] }}
Example 1. An FDO linked metadata record iin JSON-LD format. (Cut and paste into the JSON-LD Playground to visualize.)
The KO described in the linked metadata record above is available here for inspection. As Example 1 shows in bold text, our initial prototype linked metadata record for KOs relies on three vocabularies, Dublin Core Terms, the Function Ontology, and our own Knowledge Object Implementation Ontology (KOIO). As its FDO identifier, the KO uses an Archival Resource Key (ARK). ARKs are attractive because they support a suffix passthrough mechanism for consistently identifying the common parts of a KO, such as Deployment and Service Descriptions. This linked metadata record in Example 1 has been successfully loaded into several RDF systems, including the JSON-LD Playground and an instance of the Blue Brain Nexus knowledge graph system. We have used SPARQL queries to extract and filter elements from this linked metadata record.
Conclusion
For FDOs containing computable knowledge to have high-degrees of FAIRness, extensive metadata records are required. Some metadata content specified to date is specific to this type of FDO and payload. It is possible to represent FDO metadata as linked metadata, making the metadata richer semantically and potentially easier to manage with artificial agents and machines. In biomedicine especially, more work is needed to identify more vocabularies for use as controlled terminologies to arrive at suitably comprehensive linked metadata for this important new type of FDO.
Keywords
Biomedical, Biomedicine, Knowledge Grid, MCBK
Presenting author
Marisa Conte
Presented at
First International Conference on FAIR Digital Objects, presentation
Author contributions
AF authored this Abstract. All authors edited and commented on this Abstract. AF, MC, CPF, and RR collaborated on the metadata scoping work. AF and PB collaborated on representing metadata as linked metadata.
References
- Categorizing metadata to help mobilize computable biomedical knowledge.Learning Health Systems6(1). https://doi.org/10.1002/lrh2.10271
- CBK model composition using paired web services and executable functions: A demonstration for individualizing preventive services.Learning Health Systems(In Press). https://doi.org/10.1002/lhr2.10325
- Linked data on the web (LDOW2008).Proceeding of the 17th international conference on World Wide Web - WWW '08https://doi.org/10.1145/1367497.1367760
- FAIR Digital Objects for Science: From Data Pieces to Actionable Knowledge Units.Publications8(2). https://doi.org/10.3390/publications8020021
- Summary of fourth annual MCBK public meeting: Mobilizing computable biomedical knowledge-metadata and trust.Learning health systems6(1):e10301. https://doi.org/10.1002/lrh2.10301