|
Research Ideas and Outcomes : NIH Grant Proposal
|
|
Corresponding author: Arno Klein (arno@binarybottle.com)
Received: 14 Apr 2016 | Published: 22 Apr 2016
© 2016 Arno Klein.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Klein A (2016) Crowdsourcing voice editing and quality assessment of data collected from the largest mobile phone-based research study of Parkinson disease. Research Ideas and Outcomes 2: e8848. doi: 10.3897/rio.2.e8848
|

|
Executive summary
Mobile phones provide a new way of collecting behavioral medical research data at a scale never before possible – Sage Bionetworks’ mPower Parkinson research app, launched at Apple’s March 9, 2015 ResearchKit announcement, is currently collecting data related to Parkinson symptoms, such as voice recordings, from thousands of registered study participants. Before making such voice data available to any qualified researcher in the world, they need to undergo quality control and editing, which is currently something only a human can do well. To achieve this goal and the required scale, we will crowdsource these tasks through Amazon's Mechanical Turk.
Keywords
mPower, Parkinson disease, mobile health, app, voice analysis, features, crowdsourcing, annotation
Research & Related Other Project Information
Project Decsription
The principal aim of this proposal is to crowdsource the editing and quality assessment of Parkinson audio recordings collected using the mPower Parkinson mobile health research application. This application is among the first in what will become a standard way of collecting sensor-based behavioral health data from vast numbers of people, and is already collecting thousands of audio recordings, among other data. A major challenge when collecting so much data is to ensure the data quality is high. Currently, only a human can perform reliable quality assessment and editing of voice data, and the only way sufficient numbers of humans can be organized to undertake this task is by some means of crowdsourcing, such as through Amazon's Mechanical Turk.
To support this aim, we must (1) prepare the audio recordings for access via Amazon’s Mechanical Turk for thousands of people to listen to, rate, and edit the recordings, (2) provide expert (gold standard) annotations to evaluate crowdsourced results, and (3) aggregate the crowdsourced results for further analysis. For an exploratory aim, we will train a supervised learning algorithm on the crowdsourced results and evaluate how closely the automated approach matches human assessments.
Crowdsourcing mobile health research data assessment/preparation will improve data quality at a scale beyond what any research lab could possibly support. As more such research applications come into use, a successful example of crowdsourcing data assessment/preparation will encourage future collection of unstructured (audio/text/image/video) data, enriching our understanding of Parkinson disease and other conditions.
Public Health Relevance Statement
Mobile phones provide a new way of collecting behavioral medical research data at a scale never before possible – Sage Bionetworks’ mPower Parkinson research app, launched at Apple’s March 9, 2015 ResearchKit announcement, is currently collecting data related to Parkinson symptoms, such as voice recordings, from thousands of registered study participants. Before making such voice data available to any qualified researcher in the world, they need to undergo quality control and editing, which is currently something only a human can do well. To achieve this goal and the required scale, we will crowdsource these tasks through Amazon's Mechanical Turk.
Facilities & Other Resources
Laboratory: N/A
Clinical: N/A
Animal: N/A
High Performance Computing Resources: Sage Bionetworks uses a combination of scalable cloud-based storage and analytical computational resources and its own computational facilities. The cloud-based services are procured from Amazon Web services on a fee for service basis and provide a cost-effective solution to variable needs, technology upgrades and support. Sage Bionetworks develops and operates two software as a service platforms, Bridge and Synapse, as resources for the broader scientific community. Both these systems opperate on cloud-based infrastructure. Internal research projects also have access to the Sage Bionetworks high performance computing cluster, maintained through a partnership agreement with the University of Miami.
Additional servers used by Sage scientific staff are co-located at the Fred Hutchinson Cancer Research Center computing facilities. All networked file systems, databases, and home directories are backed up using Veritas software to a robotic tape library. Tapes are taken off-site each month for disaster recovery.
Additional facilities/resources :
Sage Bionetworks Bridge platform is designed to support the design and execution of adaptive clinical trials delivered though smartphone platforms. Participants in a study interact with the Bridge server through an app which is custom-designed for each particular study. The Bridge server provides a way to configure a study with a set of survey questions to ask study participants and way to schedule times for the various survey and app data collection tasks to run. Storing this configuration on the server allows researchers to dynamically adjust the study as data is collected without distributing new builds of the app to participants.
The Bridge server provides services for study participants to create an account, authenticate, and manage informed consent to participate in a research study. The app will periodically save survey and sensor measurements to the server; client-side SDKs help manage transmission of his data over intermittent mobile network connections. Study data is stored separately from readily identifying user account data so that study participants can be deidentified and data shared protecting their anonymity. Once deidentified, study data will be stored on Sage Bionetworks Synapse platform.
Sage Bionetworks Synapse platform will be used to support the complex, interrelated analyses described in this project. Synapse is an informatics platform for collaborative, data-driven science that combines the power of community-based modeling and analysis with broad access to large datasets, which will enable the development of more predictive computational models of disease. Synapse is built as a web service-based architecture in which a common set of services is accessed via different sets of client applications, including a web portal and integrations with multiple analysis environments.
Synapse is designed to allow users to bundle together and publish the relationships, annotations, and descriptions of files that may live in multiple locations. This may be files stored in Synapse own native storage location (Amazon S3), data living on local file systems, or code from GitHub. As long as the storage location is accessible via http/ftp it can be accessed through Synapse. Synapse includes analysis clients for users working in the R and Python programming languages, and a tool that runs at the program line of the Linux shell, providing basic functionality to interact with the system no matter what analysis tools the analyst would like to use. These tools allow users to query and load data, post results, and create provenance records directly from the command line. The output of any analysis can be stored in the Synapse Amazon S3, or externally and indexed in Synapse just as the underlying data. The Synapse web portal is an environment for sharing data, results, methods and tools, and is a place that enables the tracking of analysis steps and publication of analysis results to collaborators and eventually the broader community. The Synapse web portal will allow researchers to publish analysis results and track project information. A Provenance visualization tool will allow users to formally track the relationship among resources in the system and better document and communicate each analysis step.
The Synapse system is operated as a hosted service offering from Sage Bionetworks, requiring no installation of software or IT burden on the collaborating institutions. Researchers will be able to create accounts in the system and immediately use the system to collaborate.
At this time Synapse is hosted at http://synapse.org, and is being used to host a variety of bio-molecular data sets and analytical pipelines to curate this data. Sage Bionetworks has successfully used Synapse to support a number of large scale collaborative projects including open challenges in the predictive modeling of breast cancer survival and the TCGA Pan-Cancer working group.
Scientific Environment: Sage Bionetworks leases approximately 3,900 sq ft of office space on the Fred Hutchinson Cancer Research Center campus (FHCRC or the Center.) As part of the agreement with FHCRC, all Sage Bionetworks staff have full access to the Center’s research. Sage Bionetworks has a services agreement with FHCRC for facilities related logistics.
Equipment
N/A
Specific research plan
Specific aims
A new source of biomedical big data is the mobile phone medical research application. On March 9, 2015, Apple launched its ResearchKit platform for IRB-approved medical research applications to vastly scale up behavioral medical research data collection via iPhones, of which there are hundreds of millions. Our “mPower” Parkinson disease (PD) symptom tracking research study app was one of the five initial apps using ResearchKit, and in its first three months has already collected data from almost 70,000 participants. An example activity on the app is to say “Aaaaaah” into the microphone. This simple task, when performed in a laboratory, has already been shown to help classify people into those with and without PD and has shown promise at helping to estimate symptom severity of PD patients [
However, audio quality recorded under non-laboratory conditions suffers from ambient noise and other problems that current computer algorithms are unable to account for. This means that we would need human operators to perform quality control of the mPower voice recordings for these recordings to generate useful features for analysis, and with this many recordings, we would need to enlist the help of many people. And to improve upon, and in the future automate assessment, we will need to gather information about the problems in these recordings. Therefore, the primary objective of our project is to crowdsource the quality assessment and annotation of Parkinson disease voice recordings.
Aim 1: Establish gold standard data to evaluate crowdsourced results.
To evaluate the voice annotations of non-experts, we will have an expert annotate over five hundred mPower voice recordings. These annotations will indicate what problems exist in each recording, and will serve as gold standards to help us determine which non-experts are performing well, and enable us to weight their contributions accordingly.
Aim 2: Crowdsource quality assessment and annotation of Parkinson voice recordings.
To crowdsource quality assessment and annotation of mPower voice recordings, we will build a Web application to collect annotations from thousands of Amazon’s Mechanical Turk (AMT) “Workers.” Each voice recording will be presented to multiple Workers to establish inter-rater reliability statistics. Finally, we will combine these annotations to guide automated editing or processing of the recordings in preparation for audio feature extraction and selection. As in prior research conducted by Dr. Little and colleagues [
Exploratory Aim: Train a supervised learning algorithm on the crowdsourced results.
Crowdsourcing data quality control, such as proposed in this project, can solve the problem of ensuring good data quality at scales of data collection that have never before been attempted. We propose to scale this up even further by training a supervised learning algorithm on these human assessments, and evaluate how closely an automated approach matches these assessments.
This project will be the largest study ever conducted analyzing Parkinson voice data acquired over mobile phones. Such a study would simply be impossible without careful data curation, which is growing at a scale that necessitates the participation of many people. By using AMT, we will take an existing crowdsourcing solution and apply it to already gathered data to conduct our study.
Research strategy
(A) SIGNIFICANCE
How mHealth technologies can revolutionize the clinical treatment and quality of life of Parkinson disease patients: Parkinson disease (PD) is a neurological disease with a complex constellation of symptoms that currently affects over one million Americans. In spite of the disease’s complexity, the routine standard of care for monitoring the progression of PD, even in today’s age of Big Data biomedical health technologies, is still a doctor’s appointment, where the doctor administers a standard survey and assesses the patient’s physical performance via a small number of tasks. Furthermore, due to cost and inconvenience to the patient, doctors’ appointments are months apart, making it nearly impossible to objectively monitor patients’ symptom progression with any meaningful time resolution. Perhaps as a result of this uninformative regimen, it is telling and not surprising that a recent study of PD patients showed that 42% of PD patients didn’t see a neurologist once during the three-year study period (
In such a setting, we believe that distributed sensors and mobile health (mHealth) technologies such as those that PD patients could wear continuously, paired with advanced computational techniques to analyze tremendous quantities of longitudinal data, can take the neurologist’s understanding and ability to treat PD to an unprecedented level of understanding. Sensor-based technologies have the potential to create new feedback loops for patients as well as clinicians that will allow better disease management through frequent, low-cost, longitudinal tracking. Ultimately this approach could scale to clinical management in PD, and then to other brain disorders, with an impact that would revolutionize quality of care, lower costs, and our understanding of the natural history of the disease.
mHealth apps: the birth of smartphone-enabled research studies with the potential for tremendous scale: The March 9, 2015 launch of Apple’s open source ResearchKit platform for designing and conducting IRB-approved, electronically consented health research via a mobile phone heralds a new age of clinical research. Mobile phone-based clinical studies present an alternative to today’s conventional clinical studies that rely on infrequent or short-lived patient visits, such as those currently in place for PD. mHealth studies such as those enabled by ResearchKit have the potential to alter the scope of data collection since an mHealth study can enroll anyone with a smartphone wherever they happen to be, and the frequency of data collection can happen both continuously and at designated times throughout the regular day of the study participants.
The launch of Apple’s ResearchKit featured our “mPower” app as one of the first five research study apps to open for enrollment on ResearchKit. mPower is a PD symptom tracking research study app. In the first two months following its launch, over 5,000 individuals have enrolled in the mPower study and used the app to collect both self-reported survey and activity data. mPower uses a mix of standardized PD-specific surveys and tasks that activate phone sensors to collect and track health and symptoms of PD progression related to, for example, memory, dexterity, balance and gait. Our preliminary assessment of the mPower data collected during the memory, dexterity, balance and gait tasks indicates that they are of high quality and ready to serve as the basis for computational study.
In addition to these tasks, mPower also includes a task that prompts participants to say “Aaaaaah” into the phone’s microphone. With respect to this voice phonation task, our preliminary work has already demonstrated that when performed in a controlled setting, the resulting recordings from this simple activity can accurately classify people as having or not having PD. Based on these preliminary findings, we believe mPower’s voice recordings have the potential to help estimate symptom severity of a patient with PD [
However, in contrast to the high data quality of mPower’s other tasks, the voice data not surprisingly suffers from ambient noise, interruptions and other artifacts that are currently best detected and assessed by humans. Coupled to this issue is that of scale: the number of voice recordings collected in just the first three months of the mPower study approached 70,000. Thus, in order to serve as the basis of robust computational study that is maximized for statistical power, we need an approach to data cleaning that can both meet our quality objectives as well as be implemented at the scale of this rapidly growing data set.
Crowdsourcing science with Amazon’s Mechanical Turk (AMT): Amazon’s Mechanical Turk provides an online, on-demand, scalable workforce, where each “human intelligence task” (HIT) is submitted by a Requester and is performed by one of tens of thousands of paid Workers around the world. AMT is considered a convenient, affordable way to attract many workers to perform tasks that a computer is currently poor at executing. The median wage is approximately 1-2 dollars per hour, and short tasks (around 5 minutes) are awarded around 10 cents.
AMT has been evaluated for and used in many scientific studies, particularly in the social sciences [
(B) INNOVATION
Aim 1 is innovative since it will introduce the world’s first manually annotated set of phonations (audio recordings) acquired from individuals with and without PD. Such recordings (raw and processed) and meta-data (information about the quality of these recordings) can serve as a gold standard dataset in other areas of research, such as in the development of algorithms that automatically assess audio quality or edit audio data.
We also plan on using this dataset within an open challenge (see our DARPA seedling project description below). Since 2012, Sage Bionetworks has partnered with DREAM (Dialogue for Reverse Engineering Assessment and Methods) to run crowdsourced “Challenge” competitions. Founded in 2006 by IBM’s Dr. Gustavo Stolovitzky (see his Letter of Support), DREAM Challenges engage diverse communities of experts and non-experts to competitively solve a specific problem in biomedicine in a given time period. Since 2006, DREAM has launched 34 successful Challenges, published over 60 DREAM Challenge-related papers, and aggregated a “crowd” of over 8,000 solvers. DREAM’s track record of success relates to five key ingredients of its crowdsourcing model (
Rapidly make new and often unpublished data sets available for crowd-based research.
Help determine if the toughest questions in science can be solved.
Provide an objective approach for evaluating different answers to a given scientific question.
Accelerate research by virtue of crowdsourcing the data.
Build a community of experts collaborating in real time.
Aim 2 is innovative since it will be the first instance of crowdsourcing PD data processing or analysis, and will be driven by the involvement of thousands of non-patients. Indeed, the results of this project will constitute the largest study ever conducted analyzing PD voice data acquired over mobile phones. Aim 2 is innovative also because it will create a program that will enable anyone to annotate audio files within AMT. We are aware of programs that allow a Worker to listen to audio files for transcription (e.g., http://thedesignspace.net/MT2archives/001038.html#more), but don’t know of any for directly annotating or editing audio. Other researchers will be able to build on our audio annotation program once we share it through psiTurk’s Experiment Exchange (see below).
The Exploratory Aim proposes to use crowdsourced edits and ratings of audio recordings to train a machine learning algorithm to perform the same task on unedited and unrated audio recordings. If this algorithm performs well, this will be an innovative and important contribution to voice data analysis and mobile phone voice quality assessment generally, and to future PD studies in particular.
(C) APPROACH
Aim 1: Establish gold standard data to evaluate crowdsourced results.
Dr. Max Little (Aston University) is a world expert in PD audio recording and analysis, having led the Parkinson’s Voice Initiative (parkinsonsvoice.org) and having published research on estimating MDS-UPDRSS (
Dr. Little collaborated with Dr. Klein on a previous project to assess the quality of audio recordings of PD patients gathered via PatientsLikeMe.org (see DARPA seedling project description below), and on the mPower Parkinson research app project. Dr. Little will himself rate and edit at least 500 of the recordings using our Web application (see below) for use as a gold standard to evaluate each Worker’s accuracy and consistency. We will also use this information to weight Worker contributions accordingly. We will release these edited and annotated voice data alongside their original recordings as a project in Sage Bionetworks’ Synapse platform.
Aim 2: Crowdsource quality assessment and annotation of Parkinson voice recordings.
We will present 100,000 audio recordings to AMT Workers until each recording has been annotated by at least five Workers. At one cent per recording (a reasonable rate on AMT), one hundred thousand recordings annotated by five Workers each would result in a total cost of only $5,000.
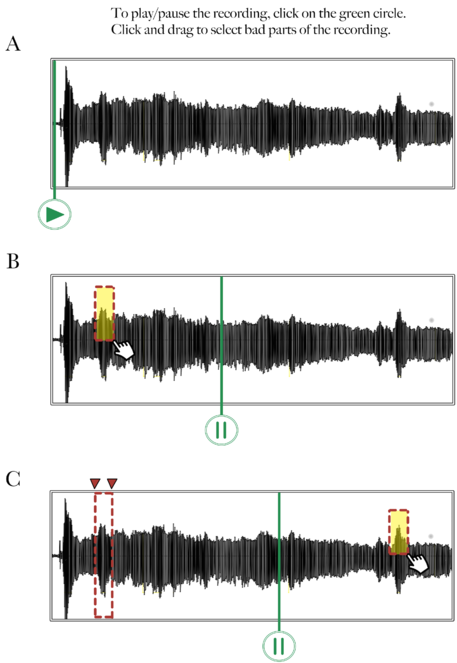
Web application: We will create a Web application accessible through AMT that will enable a Worker to edit and annotate (rate the quality of) an audio recording. A Worker will listen to a voice recording, identify problematic segments of the recording (
Mockup of audio recording annotation tool – Step 1: Selection.
This figure shows a mockup of what an audio annotation Web application tool could look like. In this first step, (A) the Worker presses the Play icon to listen to the voice recording, (B) selects a problematic segment by clicking and dragging the mouse over the waveform, and (C) replays the recording if necessary and selects other problematic segments.
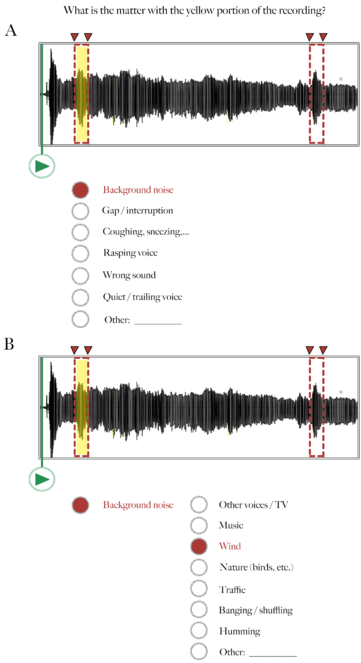
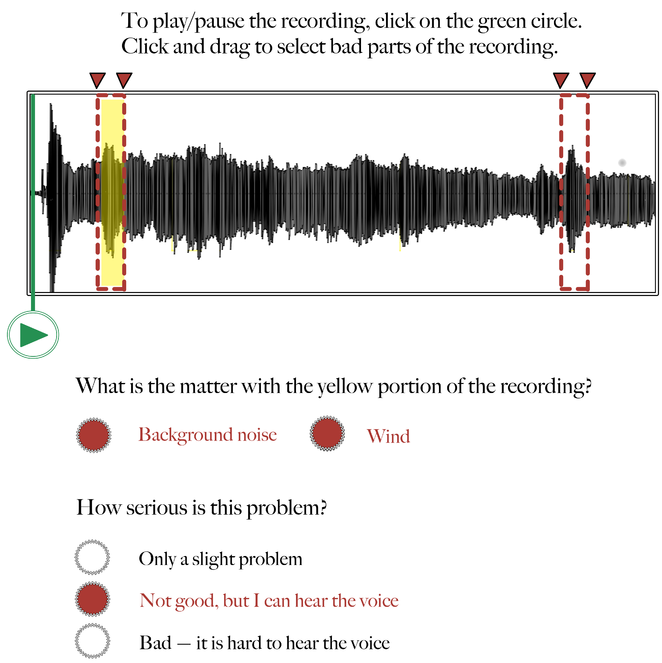
Audio recording annotation tool – Step 2: Annotation.
Following Figure 2, here the Worker selects one or more categories describing why the highlighted segment in the audio waveform is problematic. In this example, there was a lot of background noise (wind).
Audio recording annotation tool – Step 3: Rating.
Following Figures 2 and 3, here the Worker rates how serious the problem is that is affecting the highlighted segment of the recording. In this example, the Worker indicates that the background noise (wind) is not good, but that it doesn't interfere with his/her ability to hear the voice in the recording.
Training: The average person will not know how to edit a voice recording, so there will need to be some initial training and assessment. Training will be in two stages: tutorial, and evaluation. During the tutorial, a Worker will be presented with five or so recordings, each containing a problematic segment due to, for example, background noise. Each recording will have been previously edited and annotated by an expert (Dr. Little, Aim 1) but this information will not be shared with the Worker. The waveform of the recording will be visually displayed (see
Simplifying the task: Often in crowdsourcing, more complicated tasks have lower precision and inter/intra-annotator agreements. A way to handle this and collect cleaner data is to break the tasks down into smaller components (
Aggregating the data: From at least five different edits and ratings per recording, we will assess inter-rater reliability, and be able to create a consensus edit of the audio so that only the highest quality portions of each audio recording can be selected for further analysis.
Exploratory Aim: Train a supervised learning algorithm on the crowdsourced results.
We have considerable experience estimating MDS-UPDRS scores from voice data by training classifiers and regression models (see preliminary studies), such as linear regression, ridge regression, lasso, elastic net, KNN regression, random forest, boosted regression trees, etc. For this Exploratory Aim, we will use supervised machine learning methods to train on the crowdsourced annotations (start and stop points for each problematic segment of each recording, the problem category, and the severity rating). We will then evaluate how well the methods estimate MDS-UPDRS scores for a test set of voice recordings. We will use the following open source software packages: the scikit-learn and Dato Python packages, and our own software in the R programming environment.
We will design a proper semi-supervised learning model to:
- Synthesize our gold standard annotations with the AMT Workers’ annotations.
- Do supervised classification of annotated audio recordings.
- Perform unsupervised classification of audio recordings which have not been annotated.
This approach would need a custom learning algorithm. One benefit of this modeling approach is that it can be transferred to other situations with a variable amount of annotation information.
Preliminary studies:
DARPA seedling project
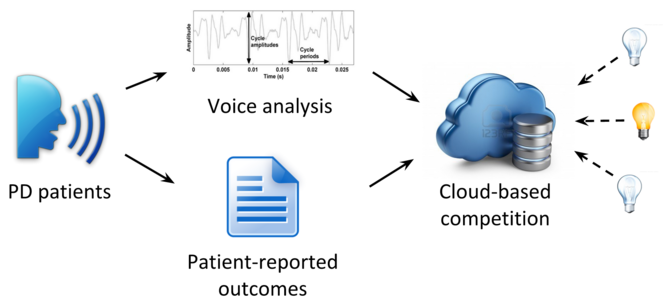
Last year, we concluded a project in collaboration with PatientsLikeMe to acquire over 600 phone recordings of phonation in PD patients to test the feasibility of conducting a voice analysis challenge (competition). Like some of our other challenges (see Innovation section), this challenge would make biomedical data (voice recordings) available to anyone in the world to challenge them to estimate a behavioral measure (MDS-UPDRS score). After crowdsourcing data collection, as in the proposed project, the challenge would constitute a second stage of crowdsourcing, of data analysis (
DARPA-funded seedling project.
This schematic represents our DARPA-funded seedling project to assess the feasibility of collecting phone voice recordings from PD patients for use in a competition.
The overall deliverables for the seedling project included:
The development of IRB-approved governance procedures for challenge-based data analysis of voice data and patient-reported outcomes gathered online,
A software-based online method to address data collection challenges,
A dataset of over 600 voice recordings and outcome data from people with PD integrated into Sage Bionetworks’ Synapse data analysis platform (http://synapse.org), and
A completed dry-run to demonstrate the feasibility of conducting a crowd-based analysis challenge, accompanied by a study report addressing the lessons learned.
It was this project that brought Dr. Klein and Dr. Little together as collaborators, and clearly exposed us to problems of gathering audio data “in the wild” (landlines from English-speaking countries in that project) and the importance of very clear instructions, a uniform platform for collecting voice data, and recording on the phone itself versus transmitting over a phone line or network.
We concluded that without proper editing and quality control of audio recordings, a crowd-based analysis challenge was not feasible. It is for this reason that we propose the current project.
Android Parkinson app
With our collaborator Dr. Ray Dorsey (PD expert at the University of Rochester), Dr. Little helped to develop and test an Android app for people with PD that records activities (phonation, finger tapping, reaction time, gait, balance) to infer PD symptom severity. In a pilot study [
Android and iOS Parkinson app screenshots.
Top: Android PD app screenshots showing instructions for the phonation (voice) task.
Bottom: mPower PD app screenshots. Each participant in the mPower study is prompted to perform a voice activity three times a day. The rightmost screenshot demonstrates the visual feedback that is provided during audio recording, to try to keep the voice at the best amplitude for recording.
mPower Parkinson iOS app
Sage Bionetworks, the University of Rochester, and Apple recently launched an iOS app that includes activities in the Android Parkinson app, passive sensor feeds, and select UPDRS survey questions. We are continuing to collect data from thousands of participants. The mPower voice recordings and UPDRS questionnaire results provide data for crowdsourcing analysis in the current project (
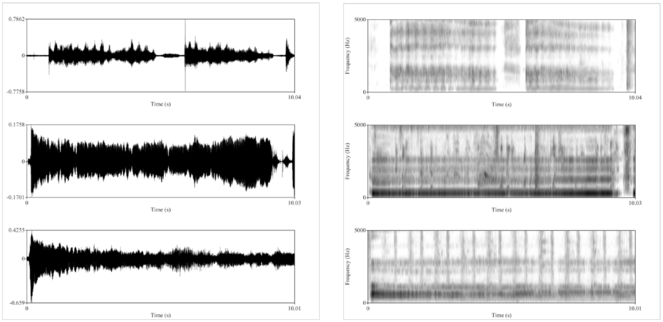
Example mPower patient voice data.
In the mPower app, PD patients are prompted to perform the voice activity three times per day: once before taking their medication, a second time when they feel they are at their best after taking their medication, and a third “random” time. This figure shows example voice data for a single patient on medication (top) and at a “random” time, very likely off medication (bottom). On the left are waveforms, showing the acoustic voice signal over time (0-10 seconds), from which one can clearly see that the patient’s voice trailed off to a minimum (bottom left) compared to after medication (top left). On the right are spectrograms, representing signal amplitude at different frequencies (0-5 kHz) over time (0-10 seconds). The spectrogram after medication (top right) has more uniform frequency bands across the recording compared to the rather “muddled” spectrogram recorded at the random time (bottom right).
Contingencies and Timeline:
Given our expertise in PD voice analysis and Web app development, we don’t anticipate a problem creating the browser-based audio annotation tool (Aim 2) or using this tool to annotate voice recordings ourselves (Aim 1). We have listened to and reviewed hundreds of recordings, and we believe that the majority of recordings will be useable after annotation and processing.
As for the crowdsourcing itself (Aim 2), many data quality projects and scientific studies have been run using AMT, but in case we have difficulties, there are a considerable number of alternatives to AMT for crowdsourcing. We should be able to attract Workers, given that we will be posting frequently and will have a large number of tasks, and it is known that Workers look for tasks that are recently posted and which have the largest number of tasks available [
Timeline.
We will prepare and update the Web app (Aim 2) as we develop it for use in annotating gold standard audio data (Aim 1) and as we get feedback on its use in connection with Amazon’s Mechanical Turk (Aim 2). Year 2 will consist primarily of testing the aggregation of annotated audio data for further analysis (Aim 2), to train an automated approach (Exploratory Aim), and to publish and present our findings.
Vertebrate animals
N/A
Resource sharing plan
Data Sharing Plan: Just as the original voice recordings collected through the mPower Parkinson mobile health research application data will be made publicly available to qualified researchers, so too will the edited, rated, and annotated versions be made available, via synapse.org.
Software Dissemination: As a 503(b) non-profit company dedicated to building support for open science, Sage Bionetworks is fully supportive of requirements to ensure the software remains a community resource.
All software developed as part of this project, such as the Web application for editing, rating, and annotating voice recordings, as well as any analysis software, will be released under the open source Apache Version 2 license and will be freely available as a public project maintained on GitHub.com and through Synapse.org.
Use of the Apache license and dissemination and maintenance of the software as a public git repository satisfy the requirements of this RFA's Resource Sharing Plan. The software will:
- be freely available,
- allow for commercialization, redistribution, and incorporation in other software,
- be maintainable by anyone thanks to git's distributed version control architecture,
- be customizable and shareable, and
- allow for pull requests to improve the software's project page on GitHub.com.
Synapse, where the data will be stored and processed, is licensed under commonly used open source licenses. The majority of the components are licensed under Apache v2 license, allowing widespread adoption. The Synapse R client is licensed under the L-GPL to be compatible with the rest of the R programming language. This ensures that the software is freely available to biomedical researchers and educators in the non-profit sector, such as institutions of education, research institutions, and government laboratories. It also ensures that outside researchers can modify the source code and to share modifications with other colleagues as well as with the investigators, while leaving commercialization opportunities available for future development. Copyright is held by Sage Bionetworks, and we will transfer these rights to another party in the event our organization ceases operations.
All Synapse source code is hosted on GitHub, allowing the community to easily create forks of our mainline development. Git Hub also has a “pull request” mechanism which is well-suited for accepting patches as contributions for outside developers. Our own developers use this mechanism to move code into the master Synapse repositories, and we’ve already successfully used the pull request mechanism to accept contributions from outside developers. Sage Bionetworks code can be found on GitHub at https://github.com/sage-bionetworks.
Additionally, our developer wiki documentation and Jira issue tracker are publically available on the web. This gives visibility into our development practices and roadmap to external developers, facilitating community contributions.
Funding program
Big Data to Knowledge (BD2K) Advancing Biomedical Science Using Crowdsourcing and Interactive Digital Media (UH2) (RFA-CA-15-006)
Project
The principal aim of this proposal is to crowdsource the editing and quality assessment of Parkinson audio recordings collected using the mPower Parkinson mobile health research application. This application is among the first in what will become a standard way of collecting sensor-based behavioral health data from vast numbers of people, and is already collecting thousands of audio recordings, among other data. A major challenge when collecting so much data is to ensure the data quality is high. Currently, only a human can perform reliable quality assessment and editing of voice data, and the only way sufficient numbers of humans can be organized to undertake this task is by some means of crowdsourcing, such as through Amazon's Mechanical Turk.
To support this aim, we must (1) prepare the audio recordings for access via Amazon’s Mechanical Turk for thousands of people to listen to, rate, and edit the recordings, (2) provide expert (gold standard) annotations to evaluate crowdsourced results, and (3) aggregate the crowdsourced results for further analysis. For an exploratory aim, we will train a supervised learning algorithm on the crowdsourced results and evaluate how closely the automated approach matches human assessments.
Crowdsourcing mobile health research data assessment/preparation will improve data quality at a scale beyond what any research lab could possibly support. As more such research applications come into use, a successful example of crowdsourcing data assessment/preparation will encourage future collection of unstructured (audio/text/image/video) data, enriching our understanding of Parkinson disease and other conditions.
Call
Big Data to Knowledge (BD2K) Advancing Biomedical Science Using Crowdsourcing and Interactive Digital Media (UH2) (RFA-CA-15-006)
Hosting institution
Sage Bionetworks
Ethics and security
Protection of Human Subjects
This research project meets the conditions for exemption under 45 CFR §46.101(b)(4), which states that the following category of research is exempt from the requirements of 45 CFR 46:
“Research, involving the collection or study of existing data, documents, records, pathological specimens, if these sources are publicly available or if the information is recorded by the investigator in such a manner that subjects cannot be identified, directly or through identifiers linked to the subjects.”
The voice data collected in the study does not represent identifiable speech recordings. Instead, it consists only of short recordings of individuals making the vowel sound 'aaah'. The current scientific consensus in speaker identification, which is the discipline concerned with the problem of identifying individuals from speech recordings, is that this voice data would fall far below the minimum data requirement in order to identify individuals (
speech data is required, that is, we must have examples of identifiable words. By contrast, the data in this study is of the meaningless voice sounds 'aaah' alone,
several minutes of example speech from each individual is required. In this study, we only have at most 30 seconds' of voice data from each individual.
As with all trials, the participants have contributed their voices and associated data on the general understanding that their data will be used to promote progress in scientific research in this area. They consented to sharing their data broadly for research purpose.
Author contributions
AK conceived of and wrote this proposal.
Conflicts of interest
None
References
-
Economic Games on the Internet: The Effect of $1 Stakes.PLoS ONE7(2):e31461. DOI: 10.1371/journal.pone.0031461
-
High-accuracy discrimination of Parkinson’s disease participants from healthy controls using smartphones.Movement Disorders28(10):e12.
-
Evaluating Online Labor Markets for Experimental Research: Amazon.com's Mechanical Turk.Political Analysis20(3):351‑368. DOI: 10.1093/pan/mpr057
-
Error driven paraphrase annotation using mechanical turk.Proceedings of the NAACL HLT 2010 Workshop on Creating Speech and Language Data with Amazon’s Mechanical Turk.
-
Fast, Cheap, and Creative: Evaluating Translation Quality Using Amazon’s Mechanical Turk.Proceedings of the Conference on Empirical Methods in Natural Language Processing. DOI: 10.3115/1699510.1699548
-
Creating speech and language data with amazon’s mechanical turk.Proceedings of the NAACL HLT 2010 Workshop on Creating Speech and Language Data with Amazon’s Mechanical Turk.
-
Nonnaïveté among Amazon Mechanical Turk workers: Consequences and solutions for behavioral researchers.Behavior Research Methods46(1):112‑130. DOI: 10.3758/s13428-013-0365-7
-
Task search in a human computation market.Proceedings of the ACM SIGKDD Workshop on Human Computation(HCOMP '10).New York.ACMDOI: DOI=10.1145/1837885.1837889
-
Evaluating Amazon's Mechanical Turk as a Tool for Experimental Behavioral Research.PLoS ONE8(3):e57410. DOI: 10.1371/journal.pone.0057410
-
Exploring normalization techniques for human judgments of machine translation adequacy collected using amazon mechanical turk.Proceedings of the NAACL HLT 2010 Workshop on Creating Speech and Language Data with Amazon’s Mechanical Turk.
-
Data Collection in a Flat World: The Strengths and Weaknesses of Mechanical Turk Samples.Journal of Behavioral Decision Making26(3):213‑224. DOI: 10.1002/bdm.1753
-
The online laboratory: conducting experiments in a real labor market.Experimental Economics14(3):399‑425. DOI: 10.1007/s10683-011-9273-9
-
Real user evaluation of spoken dialogue systems using amazon mechanical turk.Proceedings of INTERSPEECH.,11.
-
An overview of text-independent speaker recognition: From features to supervectors.Speech Communication52(1):12‑40. DOI: 10.1016/j.specom.2009.08.009
-
Crowdsourcing User Studies with Mechanical Turk.Proceedings of the SIGCHI Conference on Human Factors in Computing Systems.ACMDOI: 10.1145/1357054.1357127
-
Conducting behavioral research on Amazon’s Mechanical Turk.Behavior Research Methods44(1):1‑23. DOI: 10.3758/s13428-011-0124-6
-
How to use Mechanical Turk for Cognitive Science Research. http://cognitivesciencesociety.org/uploads/2011-t4.pdf
-
Running experiments on Amazon Mechanical Turk.Judgment and Decision Making5:411‑419.
-
Systematic evaluation of rating scales for impairment and disability in Parkinson's disease.Movement Disorders17(5):867‑876. DOI: 10.1002/mds.10248
-
Ground truth generation in medical imaging: a crowdsourcing-based iterative approach.Proceedings of the ACM multimedia 2012 workshop on Crowdsourcing for multimedia (CrowdMM '12).ACM,New YorkDOI: DOI=10.1145/2390803.2390808
-
Crowdsourced Knowledge Acquisition:.International Journal on Semantic Web and Information Systems9(3):14‑41. DOI: 10.4018/ijswis.2013070102
-
Using Mechanical Turk to Study Clinical Populations.Clinical Psychological Science1(2):213‑220. DOI: 10.1177/2167702612469015
-
Lessons from the DREAM2 Challenges.Annals of the New York Academy of Sciences1158(1):159‑195. DOI: 10.1111/j.1749-6632.2009.04497.x
-
Cooperation and contagion in web-based, networked public goods experiments.ACM SIGecom Exchanges10(2):3‑8. DOI: 10.1145/1998549.1998550
-
Neurologist care in Parkinson disease: A utilization, outcomes, and survival study.Neurology77(9):851‑857. DOI: 10.1212/wnl.0b013e31822c9123
-
Web 2.0-Based Crowdsourcing for High-Quality Gold Standard Development in Clinical Natural Language Processing.Journal of Medical Internet Research15(4):e73. DOI: 10.2196/jmir.2426