|
Research Ideas and Outcomes :
Grant Proposal
|
|
Corresponding author: Tina Heger (t.heger@wzw.tum.de), Sina Zarrieß (sina.zarriess@uni-bielefeld.de)
Received: 12 Jan 2022 | Published: 25 Jan 2022
© 2022 Tina Heger, Sina Zarrieß, Alsayed Algergawy, Jonathan Jeschke, Birgitta König-Ries
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Heger T, Zarrieß S, Algergawy A, Jeschke JM, König-Ries B (2022) INAS: Interactive Argumentation Support for the Scientific Domain of Invasion Biology. Research Ideas and Outcomes 8: e80457. https://doi.org/10.3897/rio.8.e80457
|
|
Abstract
Developing a precise argument is not an easy task. In real-world argumentation scenarios, arguments presented in texts (e.g. scientific publications) often constitute the end result of a long and tedious process. A lot of work on computational argumentation has focused on analyzing and aggregating these products of argumentation processes, i.e. argumentative texts. In this project, we adopt a complementary perspective: we aim to develop an argumentation machine that supports users during the argumentation process in a scientific context, enabling them to follow ongoing argumentation in a scientific community and to develop their own arguments. To achieve this ambitious goal, we will focus on a particular phase of the scientific argumentation process, namely the initial phase of claim or hypothesis development. According to argumentation theory, the starting point of an argument is a claim, and also data that serves as a basis for the claim. In scientific argumentation, a carefully developed and thought-through hypothesis (which we see as Toulmin's "claim'' in a scientific context) is often crucial for researchers to be able to conduct a successful study and, in the end, present a new, high-quality finding or argument. Thus, an initial hypothesis needs to be specific enough that a researcher can test it based on data, but, at the same time, it should also relate to previous general claims made in the community. We investigate how argumentation machines can (i) represent concrete and more abstract knowledge on hypotheses and their underlying concepts, (ii) model the process of hypothesis refinement, including data as a basis of refinement, and (iii) interactively support a user in developing her own hypothesis based on these resources. This project will combine methods from different disciplines: natural language processing, knowledge representation and semantic web, philosophy of science and -- as an example for a scientific domain -- invasion biology. Our starting point is an existing resource in invasion biology that organizes and relates core hypotheses in the field and associates them to meta-data for more than 1000 scientific publications, which was developed over the course of several years based on manual analysis. This network, however, is currently static (i.e. needs substantial manual curation to be extended to incorporate new claims) and, moreover, is not easily accessible for users who miss specific background and domain knowledge in invasion biology. Our goal is to develop (i) a semantic model for representing knowledge on concepts and hypotheses, such that also non-expert users can use the network; (ii) a tool that automatically computes links from publication abstracts (and data) to these hypotheses; and (iii) an interactive system that supports users in refining their initial, potentially underdeveloped hypothesis.
Keywords
argumentation in science, scientific claims, biological invasions, hypotheses, natural language processing, ontology
1 State of the art and preliminary work
Scientific claims are usually rather broad, and the empirical possibilities to test them limited. Only if broad claims are reformulated into specific hypotheses is it possible to confront them with empirical evidence (
- provide accessible summaries of domain knowledge including basic concepts and major claims as well as their refinements,
- link this semantic representation of the field to publications and data, thus allowing to tie newly posed claims to existing domain knowledge, and
- use this basis to interactively support users in optimizing their specifications and refinements of broad claims.
To date, however, research on computational argumentation machines has often focused on analyzing the – typically textual – end result of the argumentation process by, e.g., classifying or mining formulations of claims and arguments in complex scientific texts (
This project will combine methods from natural language processing (NLP), semantic web, philosophy of science and – as an example for a scientific domain – invasion biology. The following sections review relevant related research and our preliminary work in these areas.
1.1 Modeling domain knowledge and arguments
In order to make domain knowledge hidden in publications and data available to argumentation machines, both the domain of interest and arguments and related concepts need to be formally modeled. Many fields have long recognized, that a common understanding of key terms is needed. This has resulted in the development of numerous domain specific vocabularies and more formally grounded ontologies. Based on a long tradition of organising knowledge in taxonomies, biodiversity research is one of these fields with numerous good ontologies (see e.g., ENVO for environmental terms http://www.obofoundry.org/ontology/envo.html, or the plant trait ontology https://bioportal.bioontology.org/ontologies/PTO), but also less formalised but still useful vocabularies like different species check lists. Second, knowledge graphs (KGs) as formal models have gained attention. These KGs are typically focused on factual knowledge (see, e.g.
Preliminary work: Biodiversity informatics and semantic web The König-Ries group has been working on leveraging semantic web techniques to support biodiversity research for quite some time. Most of this work is so far focused on improving the FAIRness of biodiversity data. It includes work on improvement of discoverability of data by better, semantic descriptions (
1.2 Argumentation in science
Scientific texts have traditionally been an important domain for research on argumentation and, in particular, for data-driven approaches. Pioneering work by
Preliminary Work: A hierarchical hypotheses network for invasion biology The scientific study of global change and its effects on biodiversity has many facets (
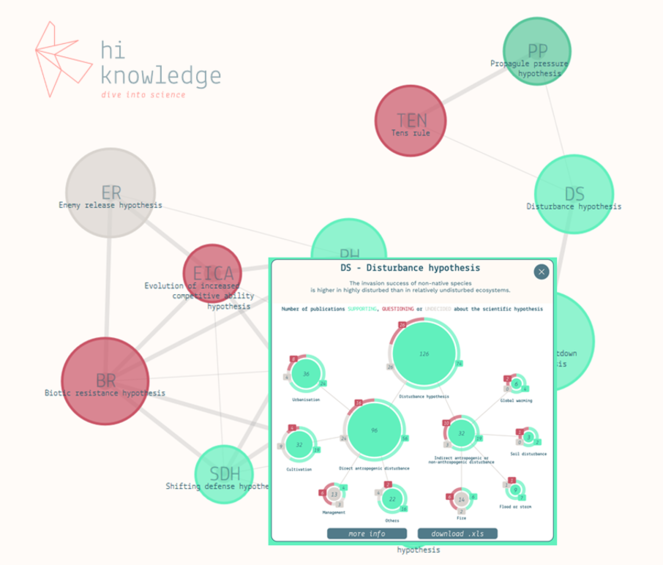
Screenshot of the website hiknowledge.org, showing a network of twelve major hypotheses on potential causes of biological invasions. The insert shows the hierarchy of hypotheses (HoH) for the disturbance hypothesis which can be retrieved by clicking on the respective dot in the network, with information on the numbers of studies supporting (green), questioning (red) or being undecided (grey) about the respective (sub)-hypotheses.
1.3 Interactive argumentation support beyond text
In NLP, argumentation support is often construed as a ‘one-shot’ classification problem, where the system’s task is to detect low-quality arguments once in a static text e.g.,
Preliminary work: Task-oriented, multi-modal dialogue A major focus of Zarrieß’ research is on task-oriented dialogue systems and interactive language generation. In
2 Objectives and work programme
2.1 Anticipated total duration of the project
36 months
2.2 Objectives
Developing a precise, new hypothesis for scientific argumentation is not an easy task. The goal of this project is to develop an interactive system that supports users in developing and refining hypotheses in invasion biology. Our interdisciplinary approach, combining methods from NLP, semantic web and philosophy of science, and drawing from in-depth domain knowledge, will combine different capabilities that users need during this process:
- domain-specific background knowledge on abstract and concrete concepts related to claims in invasion biology,
- detailed feedback on formulations of scientific hypotheses on different levels of specificity and
- links to datasets for testing hypotheses.
Fig.
We expect that our approach will be a very useful extension of HNI and contribute to the field of invasion biology, but also give general insights on how to represent knowledge for argumentation systems and leverage this knowledge for interaction with users in real-word argumentation processes. With such an approach, argumentation machines would support novice researchers in understanding the field, but would also be able to help mapping a field, detecting contradictions and gaps, and detecting links to neighboring fields, where syntactically different terms might be used to describe similar claims.
Challenges Automatic support for hypothesis development is a very challenging task for state-of-the-art argumentation machines. For research in invasion biology, the HNI in its current form is a valuable resource only for domain experts. Early career researchers and scientists new to the domain will lack background knowledge on terms, concepts (and their ambiguities) to make efficient use of the network and, e.g., find relevant abstracts. Second, scientific practice in invasion biology, and also in ecology in general, does usually not put special emphasis on precise and explicit formulation of claims or hypotheses. For example, it is usually clarified whether a claim rather amounts to the expectation of a pattern, or the suggestion of a causal relationship, or whether the claims implicitly contain unexpressed propositions. From an NLP perspective, an important challenge then is to communicate this background knowledge in an appropriate way and process potentially underdeveloped or imprecise formulations of hypotheses. Additionally, hypotheses constitute very abstract statements that, in a scientific publication, can be instantiated and formulated in very different ways. For example, two abstracts may be linked to the same hypothesis without exmplicitly mentioning it. For users not aware of certain assumptions and concepts in the field, this will be hard to determine.
These phenomena also create challenges for semantic web systems: Beyond the need for integration across domains, an approach is needed in INAS to support smooth, continuous evolution of the semantic backbone as modeling and understanding of the domain deepens and evolves. A second challenge in INAS will be the seamless integration of data as basis for arguments. This requires first of all to semantically describe data. Due to the large volume of available data, this task clearly needs to be automated. This requirement has recently sparked the SemTab challenge (http://www.cs.ox.ac.uk/isg/challenges/sem-tab/). Second, an abstraction layer needs to be added to the data turning it into an argument. This requires summarization and interpretation of data.
2.3 Work programme including proposed research methods
To address the challenges discussed above, this project brings together experts from the fields of NLP, biology and semantic web. This broad expertise will be supplemented by collaborations with philosophers of science. We believe that this is an ideal set-up to advance the state-of-the-art in argument modeling and move towards systems that meet the complex information needs of users and are flexible enough to be automatically extended to new hypotheses, new publications, new datasets and, ultimately, also new domains and other research areas.
2.3.1 Methods
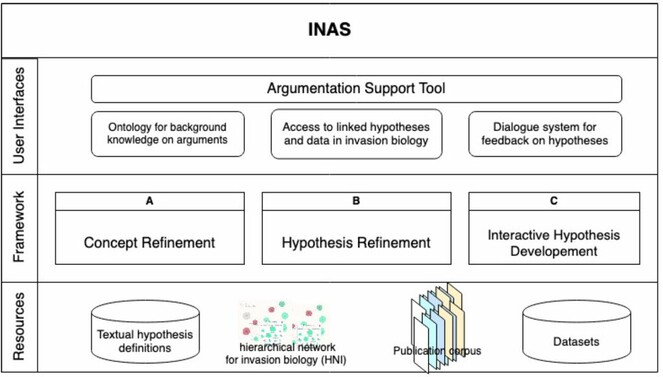
Knowledge representation Our framework will model and represent the internal semantic structure of claims in terms of abstract domain-specific concepts and their various possible refinements in testable hypotheses, as sketched in Fig.
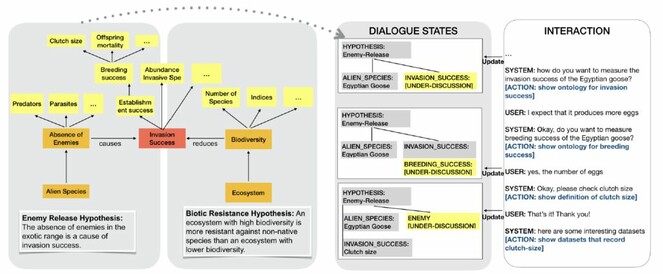
Interactive hypothesis development, based on a semantic model of hypotheses in the invasion biology domain (left) and a made-up example of a short interaction with an information-state-based dialogue system that iteratively refines a hypothesis introducing domain-specific terms in collaboration with the user (right, resolved questions appear in grey, questions under discussion in yellow).
Argumentation and data Our work will integrate multiple ways and dimensions of modeling hypotheses, i.e., in text but also in knowledge representations and through datasets. As illustrated in Fig.
Dialogue modeling We propose to model hypothesis development in a dialogue system that uses the HNI ontology to compute hierarchical information states (e.g. the general claim, concepts represented in the claim, sub-parts of the given claim) which need to be filled throughout the interaction between user and system. Thus, the system will not need to process or validate an entire argument at once, but rather focus on specifying different parts of the claim in a step-by-step, collaborative fashion, as illustrated in Fig.
Evaluation To date, there are few systematic insights into how argumentation systems should be set up to really enhance the way users can understand and develop arguments. An important goal of the project is to develop an evaluation scenario and a user study design that fills this gap and, ideally, can be generalized to other domains or other argumentation support scenarios. We plan to collaborate with other RATIO projects on this topic, e.g. with Philipp Cimiano’s and Ulf Leser’s planned project on argumentation support in a clinical domain.
2.3.2 Work packages
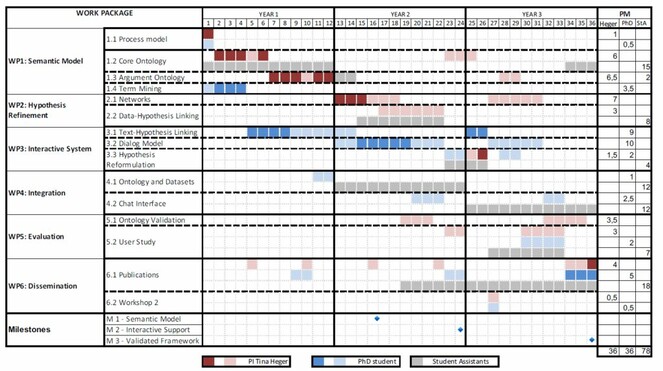
An outline of the work packages with effort in person months is given in Fig.
Milestones The project will be structured by 3 milestones (see Fig.
- M1: the basic framework for semantic modeling of hypotheses is set up
- M2: a proof-of-concept system for interactive hypothesis development is set up
- M3: the framework is integrated, validated and tested in user studies
WP 1: A semantic model for argumentation in invasion biology
A prerequisite to leveraging the power of Semantic Web techniques are shared ontologies to facilitate the seamless exchange of information. In this WP, we will bring together domain experts, philosophers of science, knowledge engineers, and end users to create such ontologies for our domain of interest (WP 1.2) and the argumentation domain and linking the two (WP 1.3). We will support this with text mining to identify key concepts, their definitions and relations (WP 1.4). Creating ontologies is not a one-time task, but rather an iterative community process which requires support for an evolving and deepening understanding of the domain (WP 1.1).
WP 1.1: Process model It is a characteristic of science that the understanding of a field becomes more nuanced over time. For us, this implies, that the domain model will also evolve over time. At the very beginning of the project, taking into account existing work on ontology evolution (
WP 1.2: Core ontology The core ontology for invasion biology, called HoH ontology, will be used to model the complex structure of knowledge in the hierarchy of hypotheses in the domain of invasion biology. We will adopt the fusion/merge strategy (
WP 1.3: Argumentation ontology Our argumentation ontology will be based on the AIF (Argument Interchange Format,
WP 1.4 : Term mining The goal of this WP is to semi-automatically obtain lists of names or terms referring to instances of species and locations, and potentially other entity types identified in WP 1.2 from the INAS abstracts. These will contribute to populating the invasion biology core ontology (WP 1.2) and to fine-tune tools for NER and argument linking in WP 3. Based on resources like LINNAEUS (
- BioBERT, a neural transformer-based network that learns word embeddings on large amounts of text from the biomedical domain and fine-tunes them for different tasks, including NER on LINNAEUS and Species-800 and
- the LSTM-CRF by
Lample et al. (2016) .
A subset of the automatic annotations obtained from BioBERT and LSTM-CRF will be corrected manually during the ontology development. These can, in turn, be used to fine-tune Bio-BERT to predict species and locations on the INAS abstracts.
WP 2: Hypothesis refinement
While the ontology development in WP1 focuses on the identification and refinement of concepts used in hypotheses in invasion biology, this work package investigates the refinement of the hypotheses themselves. We design a more detailed, nested representation of the hypotheses in the HNI (WP 2.1) and link this to datasets (WP 2.2). Fig.
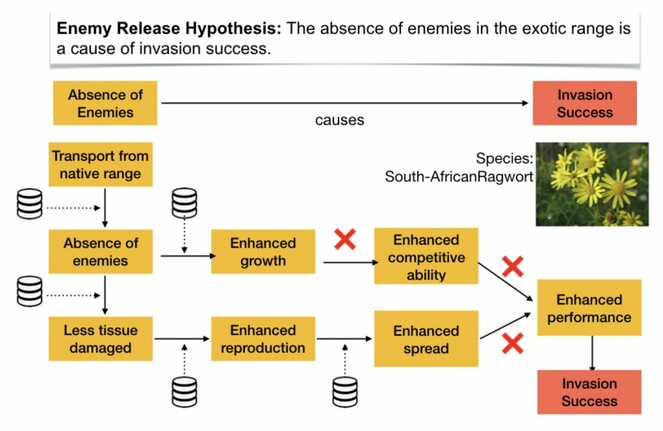
WP 2.1: Hypotheses as nested causal networks In the invasion biology domain, hypotheses often are formulated as if they would address simple causal relationships (e.g. ”The absence of enemies in the exotic range is a cause of invasion success”). For domain experts, however, these simplifications are hints to basic knowledge about underlying mechanisms, i.e. longer chains or networks of hypothesized causal relationships. In this work package, we will re-formulate the hypotheses contained in the hierarchical hypothesis network as complex, nested causal relationships. For each element in the causal chains, key references from the domain literature will be searched. The nested representations of hypotheses will be used to annotate a subset of 50-100 publication abstracts in our collection. These annotations can be used as a fine-grained test set for the NLP system in WP 3.1 and will be made available as a corpus to the NLP community (see data management plan). To fulfill this task, we will closely cooperate with philosophers of science.
WP 2.2: Data-hypothesis linking In biology, data is an important dimension of argumentation, as it is needed to test hypotheses and to support or refute claims. Detailed information on available datasets is also very important during the hypothesis development process, e.g., for exploring whether and how a certain claim has been tested in prior work (see Fig.
WP 3: Interactive Support for Hypothesis Development
In this WP, we will build an interactive system that uses the resources for concept and hypothesis refinement in WP 1 and WP 2 to support users in developing a hypothesis in the field of invasion biology. The main novelty and central challenge here is that hypothesis development is a very abstract task where communicative success is difficult to measure. We will build a neural, non-interactive classification model for text-hypothesis understanding (i.e. linking) (WP 3.1) and integrate this with a dialogue system with a predefined action-state space (WP 3.2.), which will be fine-tuned after an initial user study (WP 3.3).
WP 3.1: Text-hypothesis linking An important task of the dialogue system is to determine which general claim or hypothesis the user is talking about. We operationalize this as a classification problem, where the task is to predict whether a sentence entered by a user refers to a hypothesis represented in HNI. We will set up a neural architecture with two encoders, e.g. RNNs that learn hidden representations of the HNI hypothesis and the textual hypothesis. The central research question here is whether we can successfully leverage the symbolic knowledge encoded in the ontology (WP 1) in the neural encoders for the text and hypotheses, e.g. through compositional neural models (
WP 3.2: Dialogue model We set up a dialogue component for hypothesis development that splits up this process into a sequence of smaller steps, like e.g. discuss
- the general claim,
- the species,
- how to refine concepts in the general claim, etc., extending Zarrieß' previous work on establishing references in installments (
Zarrieß and Schlangen 2016 ).
Once the system and the user have agreed on a general claim, the subsequent states will depend on the hypothesis components represented in the ontology (WP 1), see Fig.
WP 3.3: Hypothesis reformulation As a first evaluation of the dialogue system (WP 3.2.), we carry out a pilot human evaluation with students from the biology programme in Berlin or Jena. This study will give us very valuable data on how users reformulate their hypotheses based on feedback of our system and we will use it to conduct a careful analysis of interaction quality in general and the process of hypothesis development in particular. In case we find that the interactions between our system and users are already of good quality and enable users to develop their own hypotheses, we can use the data to fine-tune/learn aspects of the dialogue system’s action space in WP 3.2 (e.g. when to give certain types of verbal or non-verbal feedback). In the other case, the data will be extremely useful to further develop our system and gain a deeper qualitative understanding of how the system can support the very challenging task of hypothesis development.
WP 4: Resources and Integration
The current version of HNI is available on the public website hi-knowledge.org. We extend this interface and integrate it with the models and resources developed in WP 1.3. The extended interface will be used to run user studies and evaluations, as described in WP 5.
WP 4.1: Ontology and datasets We will integrate the ontology from WP 1 such that users can inspect the meanings of terms used in a hypothesis description or a paper abstract. We will set up a database that records the available meta data for the papers represented in HNI, including the paper abstracts which will be indexed to support basic keyword search and links to available data sets as well as their semantic enrichment where applicable.
WP 4.2: Chat interface We will extend HNI with a simple chat interface to integrate the dialogue system from WP 3, using our web-based SLURK tool (
WP 5: Evaluation
One of the central goals of INAS is to build a framework for argument modeling that is closely tied to the needs of human users. We will thoroughly validate and consolidate the ontologies developed in WP1 with experts and conduct user studies, assessing to what extent our systems helps users in hypothesis development.
WP 5.1: Ontology consolidation and validation During a three-day workshop, the core ontology, the argumentation ontology as well as the nested representation of hypotheses will be validated. We will invite domain experts from the invasion biology community and philosophers of science. We will use a combination of pre-workshop tasks, panel presentations, break-out discussions and panel discussions to reach a broad consensus on the main features of the ontologies and the nested hypotheses. The workshop results will be used to consolidate our models.
WP 5.2: User study We will design and conduct a user study to assess the quality of argumentation support system. This includes the definition of a concrete hypothesis development task that users will have to carry out when interacting with our system (e.g. based on a given paper in invasion biology, define a promising hypothesis for follow-up studies), the identification of a target user group and the definition of criteria for assessing hypotheses that users develop with the help of our system. As users might interact very differently with our system depending on their background, we will need to identify two relatively consistent user groups (e.g. undergraduate or graduate students in biology that have taken classes on ecology) to obtain meaningful results. We will conduct a pilot user study with approx. 30 participants towards the end of the second year of the project (Fig.
WP 6: Dissemination
WP 6.1: Conferences and publications The PhD student and PI Tina Heger will present project results at international conferences and workshops. The events will cover the fields of NLP, semantic web, philosophy of science, invasion biology and ecology. The project team will publish at least 4 publications in international journals and high-ranked conferences from the fields of NLP, semantic web, philosophy of science and invasion biology as outlets. We view research data management and in particular the sustainable provision and publication of FAIR data as another important dissemination activity that will be tackled in this WP.
WP 6.2: Workshop ”Modelling the argumentation process across domains” A further element of this work package will be a workshop bringing together research groups working on similar tools in different domains. Aims of the workshop will be:
- to present our results in order to allow for exchange and synergies with related projects, and
- to compare argumentation processes and ways to model them across domains.
Acknowledgements
We thank the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) for funding this project (Project number 455913229). The publication of this article was funded by the Open Access Fund of the Leibniz Association.
Funding program
Schwerpunktprogramm "Robust Argumentation Machines (RATIO)"
Grant title
INAS: Interactive argumentation support for the scientific domain of invasion biology; project number 455913229
Hosting institutions
- University of Bielefeld
- Leibniz Institute of Freshwater Ecology and Inland Fisheries (IGB)
- University of Jena
Author contributions
Tina Heger and Sina Zarrieß contributed equally to this proposal.
References
- Towards transforming tabular datasets into knowledge graphs.The Semantic Web: ESWC 2020 Satellite Eventspp. 217‑288. https://doi.org/10.1007/978-3-030-62327-2_37
- OAPT: A Tool for Ontology Analysis and Partitioning. Demo Paper.EDBT.644–647pp.
- Towards a Core Ontology for Hierarchies of Hypotheses in Invasion Biology.The Semantic Web: ESWC 2020 Satellite Events3‑8. https://doi.org/10.1007/978-3-030-62327-2_1
- Neural Module Networks.2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR)https://doi.org/10.1109/cvpr.2016.12
- Towards a Knowledge Graph for Science.Proceedings of the 8th International Conference on Web Intelligence, Mining and Semanticshttps://doi.org/10.1145/3227609.3227689
- The ReCAP Project.Datenbank-Spektrum20(2):93‑98. https://doi.org/10.1007/s13222-020-00340-0
- A proposed unified framework for biological invasions.Trends in Ecology & Evolution26(7):333‑339. https://doi.org/10.1016/j.tree.2011.03.023
- How conversation is shaped by visual and spoken evidence. In:Approaches to studying world-situated language use: Bridging the language-as-product and language-as-action traditions.Cambridge, MA: MIT Press.,pp. 95-129pp.
- Towards an argument interchange format.The Knowledge Engineering Review21(4):293‑316. https://doi.org/10.1017/s0269888906001044
- Using Language.Cambridge University Press,Cambridge. https://doi.org/10.1017/CBO9780511620539
- What is the Essence of a Claim? Cross-Domain Claim Identification.Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processinghttps://doi.org/10.18653/v1/d17-1218
- Selecting and Tailoring Ontologies with JOYCE.Lecture Notes in Computer Science114‑118. https://doi.org/10.1007/978-3-319-58694-6_12
- Argumentative Zoning Applied to Critiquing Novices’ Scientific Abstracts.The Information Retrieval Series233‑246. https://doi.org/10.1007/1-4020-4102-0_18
- On the discoursive structure of computer graphics research papers.Proceedings of The 9th Linguistic Annotation Workshophttps://doi.org/10.3115/v1/w15-1605
- Scientific method for ecological research.Cambridge University Press,Cambridge. https://doi.org/10.1017/CBO9780511612558
- Language efficiency and visual technology: Minimizing collaborative effort with visual information.Journal of language and social psychology23(4):491‑517. https://doi.org/10.1177/0261927X04269589
- LINNAEUS: A species name identification system for biomedical literature.BMC Bioinformatics11(1). https://doi.org/10.1186/1471-2105-11-85
- Conceptual Frameworks and Methods for Advancing Invasion Ecology.AMBIO42(5):527‑540. https://doi.org/10.1007/s13280-012-0379-x
- Towards an Integrative, Eco-Evolutionary Understanding of Ecological Novelty: Studying and Communicating Interlinked Effects of Global Change.BioScience69(11):888‑899. https://doi.org/10.1093/biosci/biz095
- The Hierarchy-of-Hypotheses approach: A synthesis method for enhancing theory development in ecology and evolution.BioScience71(4):337‑349. https://doi.org/10.1093/biosci/biaa130
- ROBOT: A Tool for Automating Ontology Workflows.BMC Bioinformatics20(1). https://doi.org/10.1186/s12859-019-3002-3
- Support for major hypotheses in invasion biology is uneven and declining.NeoBiota14:1‑20. https://doi.org/10.3897/neobiota.14.3435
- Invasion biology: hypotheses and evidence.CABIhttps://doi.org/10.1079/9781780647647.0000
- Integrated Semantic Search on Structured and Unstructured Data in the ADOnIS System.S4BioDiv@ ISWC.
- Semantic technologies for consolidating structured data and unstructured documents in biodiversity research.Geoinformationssysteme 2015. Beiträge zur 2. Münchner GI-Runde.Wichmann
- Neural Architectures for Named Entity Recognition.Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies260‑270. https://doi.org/10.18653/v1/n16-1030
- An Argument-Annotated Corpus of Scientific Publications.Proceedings of the 5th Workshop on Argument Mininghttps://doi.org/10.18653/v1/w18-5206
- Context-independent claim detection for argument mining.Proc. of IJCAI.
- Confirmation of ecological and evolutionary models.Biology & Philosophy2(3):277‑293. https://doi.org/10.1007/bf00128834
- Tag Me If You Can! Semantic Annotation of Biodiversity Metadata with the QEMP Corpus and the BiodivTagger.Proc of LREC. Marseille, France: European Language Resources Association, May 2020, pp. 4557–4564.
- Dataset search in biodiversity research: Do metadata in data repositories reflect scholarly information needs?PLOS ONE16(3). https://doi.org/10.1371/journal.pone.0246099
- An Additional Future for Psychological Science.Perspectives on Psychological Science8(4):414‑423. https://doi.org/10.1177/1745691613491270
- The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text.PLoS ONE8(6). https://doi.org/10.1371/journal.pone.0065390
- Towards a biodiversity knowledge graph.Research Ideas and Outcomes2https://doi.org/10.3897/rio.2.e8767
- Essential Annotation Schema for Ecology (EASE)—A framework supporting the efficient data annotation and faceted navigation in ecology.PLOS ONE12(10). https://doi.org/10.1371/journal.pone.0186170
- Ontologies: How can They be Built?Knowledge and Information Systems6(4):441‑464. https://doi.org/10.1007/s10115-003-0138-1
- The Argument Interchange Format.Argumentation in Artificial Intelligence383‑402. https://doi.org/10.1007/978-0-387-98197-0_19
- Argumentation schemes in argument-as-process and argument as- product.Proc. of the Conference Celebrating Informal Logic.
- Argument Diagramming: The Araucaria Project.Advanced Information and Knowledge Processing164‑181. https://doi.org/10.1007/978-1-84800-149-7_8
- Training and hackathon on building biodiversity knowledge graphs.Research Ideas and Outcome5:e36152. https://doi.org/10.3897/rio.5.e36152
- Introduction to the CoNLL-2003 shared task: Language independent named entity recognition.arXiv preprintURL: https://arxiv.org/abs/cs/0306050
- slurk – A Lightweight Interaction Server For Dialogue Experiments and Data Collection.Proc. of AixDial / SEMdial. Aix-en-Provence, France.
- Towards an ecological trait‐data standard.Methods in Ecology and Evolution10(12):2006‑2019. https://doi.org/10.1111/2041-210x.13288
- Towards Knowledge Graph Construction using Semantic Data Mining.Proceedings of the 21st International Conference on Information Integration and Web-based Applications & Serviceshttps://doi.org/10.1145/3366030.3366035
- Recognizing Insufficiently Supported Arguments in Argumentative Essays.Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Volume 1, Long Papershttps://doi.org/10.18653/v1/e17-1092
- Argumentative zoning: Information extraction from scientific text. PhD thesis.University of Edinburgh
- The uses of argument.Cambridge University Presshttps://doi.org/10.1017/CBO9780511840005
- Argdf: Arguments on the semantic web’.The British University in Dubai Jointly with The University of Edinburgh
- Ontology-Based Approach to Organizing the Support for the Analysis of Argumentation in Popular Science Discourse.Communications in Computer and Information Science348‑362. https://doi.org/10.1007/978-3-030-30763-9_29
- Easy Things First: Installments Improve Referring Expression Generation for Objects in Photographs.Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)https://doi.org/10.18653/v1/p16-1058
- PentoRef: A corpus of spoken references in task-oriented dialogues.Proc. of LREC.
- Obtaining referential word meanings from visual and distributional information: Experiments on object naming.Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)https://doi.org/10.18653/v1/p17-1023
- Know What You Don’t Know: Modeling a Pragmatic Speaker that Refers to Objects of Unknown Categories.Proc. of the 57th Annual Meeting of the ACL. Florence, Italy: Association for Computational Linguistics, July 2019, pp. 654–659.