|
Research Ideas and Outcomes :
Short Communication
|
|
Corresponding author: Dao-Yin Dong (dongdy@haust.edu.cn)
Received: 04 Jun 2021 | Published: 08 Jun 2021
© 2021 Dao-Yin Dong, Pu-Yu Li
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Dong D-Y, Li P-Y (2021) Identifying SOX2-OT transcript that is responsible for regulating SOX2 in cancer cells and embryonic stem cells. Research Ideas and Outcomes 7: e69726. https://doi.org/10.3897/rio.7.e69726
|
|
Abstract
SOX2 overlapping transcript (SOX2-OT) is an evolutionarily conserved long non-coding RNA (lncRNA) whose intronic region contains the transcript of pluripotency gene SRY-box transcription factor 2 (SOX2). It has been suggested that SOX2-OT can regulate its overlapping gene, SOX2. Studies demonstrated that elevated SOX2-OT promotes SOX2 expression in cancer cells, whereas levels of SOX2-OT are inversely correlated with levels of SOX2 in embryonic stem cells. It is not clear why there is a tremendous discrepancy in the regulation of SOX2 by SOX2-OT in cancer cells and embryonic stem cells. Due to the diversified transcription of the SOX2-OT gene, we hypothesize that differential expression of transcripts of the SOX2-OT gene in cancer cells and embryonic stem cells may contribute to the divergence in the regulatory relationship of SOX2-OT and SOX2. A CRISPR screening platform can be leveraged to systemic evaluate which transcript of the SOX2-OT gene may be responsible for upregulation or downregulation of SOX2 in cancer cells and embryonic stem cells, respectively.
Keywords
SOX2-OT, SOX2, Cancer cells, embryonic stem cells, CRISPR
Differential regulation of SOX2 by SOX2-OT in cancer cells and embryonic stem cells
SOX2 overlapping transcript (SOX2-OT) is an evolutionarily conserved long non-coding RNA (lncRNA) whose intronic region contains the transcript of pluripotency gene SRY-box transcription factor 2 (SOX2) (
The human SOX2-OT gene is comprised of many exons and has multiple transcription start sites (TSSs) that exhibit complicated transcriptional features. Six RefSeq mRNAs with 15 additional mRNAs sequence are collected in Genebank (NCBI). According to Ensembl Genome Database, the human SOX2-OT gene expresses 104 mRNA-like transcripts, the longest of which is approximately 4.3 kb. In addition, RNAcentral (https://rnacentral.org/) and LNCipedia (https://lncipedia.org/) include 167 and 127 SOX2-OT transcripts, respectively. The diversified transcription increases the complexity of the SOX2-OT gene. We speculate that differential expression of transcripts of the SOX2-OT gene in cancer cells and embryonic stem cells may contribute to the divergence in the regulatory relationship of SOX2-OT and SOX2. Although some studies revealed the exact transcript of SOX2-OT can control SOX2 expression, those experiments generally studied how SOX2-OT regulates SOX2 expression by siRNA silencing (
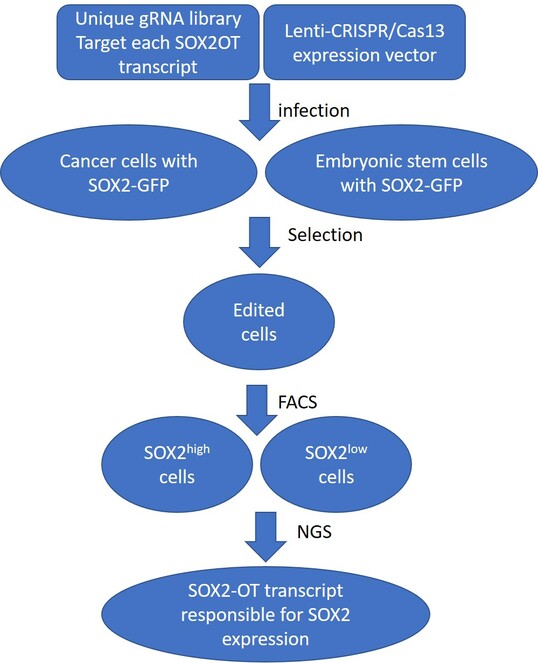
A method to identify SOX2-OT transcript that is responsible for regulating SOX2 in cancer cells and embryonic stem cells
We can deploy a platform that leverages a CRISPR screening to systemic evaluate which transcript of the SOX2-OT gene may be responsible for upregulation or downregulation of SOX2 in cancer cells and embryonic stem cells, respectively. We need to avoid the use of CRSPR/Cas9 system because genomic editing to disturb the expression of SOX2-OT may affect the expression of SOX2. SOX2 is embedded in an intron of SOX2-OT and the regulatory elements to control SOX2 expression may localize in the gene body of SOX2-OT.
CRISPR/Cas13 system has been engineered to induce RNA knockdown without genomic modifications (
Acknowledgements
This research was funded by the National Natural Science Foundation of China (grant number: 81901485)
Conflicts of interest
The authors declare no conflicts of interest.
References
- C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector.Science353(6299). https://doi.org/10.1126/science.aaf5573
- RNA targeting with CRISPR-Cas13.Nature550(7675):280‑284. https://doi.org/10.1038/nature24049
- Complex architecture and regulated expression of the Sox2ot locus during vertebrate development.RNA15(11):2013‑27. https://doi.org/10.1261/rna.1705309
- Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer.PLoS One9(7). https://doi.org/10.1371/journal.pone.0102140
- LncRNA SOX2OT affects cervical cancer cell growth, migration and invasion by regulating SOX2.Cell Cycle19(11):1391‑1403. https://doi.org/10.1080/15384101.2020.1750812
- MTA3 Represses Cancer Stemness by Targeting the SOX2OT/SOX2 Axis.iScience22:353‑368. https://doi.org/10.1016/j.isci.2019.11.009
- Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection.Nature538(7624):270‑273. https://doi.org/10.1038/nature19802
- A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival.Int J Biochem Cell Biol53:380‑8. https://doi.org/10.1016/j.biocel.2014.06.004
- SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas.PLoS One5(1). https://doi.org/10.1371/journal.pone.0008960
- Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2.Cell Death Dis9(8). https://doi.org/10.1038/s41419-018-0840-2
- Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors.Cell173(3):665‑676. https://doi.org/10.1016/j.cell.2018.02.033
- Functional Classification and Experimental Dissection of Long Noncoding RNAs.Cell172(3):393‑407. https://doi.org/10.1016/j.cell.2018.01.011
- Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma.Oncogene37(28):3822‑3838. https://doi.org/10.1038/s41388-018-0237-9
- The multidimensional mechanisms of long noncoding RNA function.Genome Biol18(1). https://doi.org/10.1186/s13059-017-1348-2
- Allele-specific repression of Sox2 through the long non-coding RNA Sox2ot.Sci Rep8(1). https://doi.org/10.1038/s41598-017-18649-4
- Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma.Stem Cells32(1):126‑34. https://doi.org/10.1002/stem.1542
- Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis.Front Genet6https://doi.org/10.3389/fgene.2015.00196
- Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28.Mol Cell65(4):618‑630. https://doi.org/10.1016/j.molcel.2016.12.023
- SOX2OT, a novel tumor-related long non-coding RNA.Biomed Pharmacother123https://doi.org/10.1016/j.biopha.2019.109725
- LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2.IUBMB Life69(11):867‑876. https://doi.org/10.1002/iub.1681
- IRF4-induced upregulation of lncRNA SOX2-OT promotes cell proliferation and metastasis in cholangiocarcinoma by regulating SOX2 and PI3K/AKT signaling.Eur Rev Med Pharmacol Sci22(23):8169‑8178. https://doi.org/10.26355/eurrev_201812_16509
- Massively parallel Cas13 screens reveal principles for guide RNA design.Nature Biotechnologyhttps://doi.org/10.1038/s41587-020-0456-9
- Yin Yang-1 suppresses pancreatic ductal adenocarcinoma cell proliferation and tumor growth by regulating SOX2OT-SOX2 axis.Cancer Lett408:144‑154. https://doi.org/10.1016/j.canlet.2017.08.032
- Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2.Mol Cancer19(1). https://doi.org/10.1186/s12943-020-1143-7