|
Research Ideas and Outcomes :
Review Article
|
|
Corresponding author: Laurence Livermore (l.livermore@nhm.ac.uk)
Received: 13 Aug 2020 | Published: 14 Aug 2020
© 2020 Stephanie Walton, Laurence Livermore, Olaf Bánki, Robert Cubey, Robyn Drinkwater, Markus Englund, Carole Goble, Quentin Groom, Christopher Kermorvant, Isabel Rey, Celia Santos, Ben Scott, Alan Williams, Zhengzhe Wu
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Walton S, Livermore L, Bánki O, Cubey RWN, Drinkwater R, Englund M, Goble C, Groom Q, Kermorvant C, Rey I, Santos CM, Scott B, Williams AR, Wu Z (2020) Landscape Analysis for the Specimen Data Refinery. Research Ideas and Outcomes 6: e57602. https://doi.org/10.3897/rio.6.e57602
|
|
Abstract
This report reviews the current state-of-the-art applied approaches on automated tools, services and workflows for extracting information from images of natural history specimens and their labels. We consider the potential for repurposing existing tools, including workflow management systems; and areas where more development is required. This paper was written as part of the SYNTHESYS+ project for software development teams and informatics teams working on new software-based approaches to improve mass digitisation of natural history specimens.
Keywords
machine learning, natural language processing, natural history specimens, data refinery, data reconciliation, semantic segmentation, digitisation, linked open data, workflow management, collections digitisation
1. Introduction
A key limiting factor in organising and using information from global natural history specimens is making that information structured and computable. As of 2020 at least 85% of available specimen information currently resides on labels attached to specimens or in physical registers and is not digitised or publicly available*
The objective of the Specimen Data Refinery (SDR) is to combine these technologies into a cloud-based platform for processing specimen images and their labels en masse in order to extract essential data efficiently and effectively, according to standard best practices.
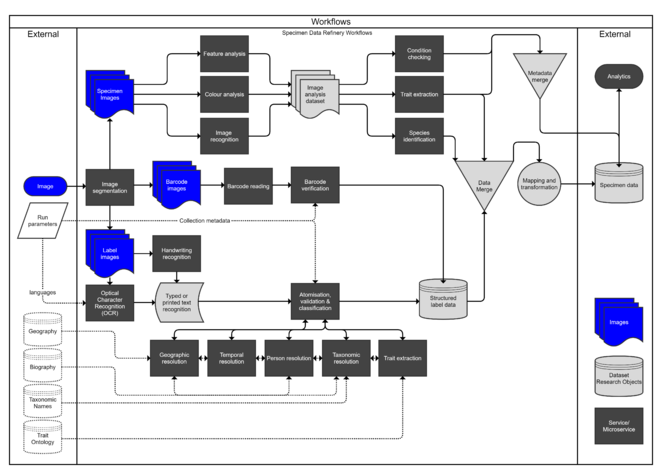
As part of this process a workflow was developed, illustrating the steps required to fully automate the procedure from image capture to a full specimen dataset (Fig.
A glossary of terms is provided at the end of this report to assist the reader on unfamiliar or specialised terminology (see Glossary).
A gap analysis was conducted, taking into account the maturity of the available tools for each phase in the workflow (Section 3).
The second component of building an automated workflow is developing the links between each tool - the environment in which the entire process is executed and the technology that executes the process. This is a different set of platforms and services that will connect what are currently various disparate pieces into a whole working system. It requires a technology stack that is reliable, sustainable and cost-effective. Following the gap analysis on tools, an initial assessment was conducted on the technology stack required to assemble these tools together into an automated workflow (Section 4).
1.1 Scope
The scope of this work was to evaluate existing platforms based on their approach and service offering, and to identify sources of data including reference/ground truth/training datasets. This analysis also identified missing tools/service and datasets.
This report does not include: technical evaluation of existing tools, service registries and platform-based approaches; evaluation and recommendations on using, integrating and merging partial (prior/previously created) specimen data; assessment of hardware and physical infrastructure requirements; assessment for the potential to use pan-European Collaborative Data Infrastructure; creation of reference/ground truth/training datasets.
1.1.1 Machine Learning and Training Data Sets
The tools evaluated in this landscape analysis include both unsupervised and supervised machine learning approaches, with a key difference being that unsupervised methods do not require a training dataset. For example, some image segmentation tools are unsupervised - their methods for identifying parts of an image include thresholding, contouring, clustering, etc and how these are applied to segment an image does not change, regardless of the number of images processed. In comparison, other segmentation approaches like U-Nets use supervised learning and require a ‘training period’ for image recognition when they are ‘taught’ to identify specific items in an image based on a ground-truth set of images (
Many of the machine learning tools included in this study are specific to natural history collections and, in many cases, are designed for specific taxa. Thus, these tools have been trained and tested with species datasets. In order to gain a comprehensive picture of the tools available, software not designed specifically for scientific collections or with limited testing on natural history collections was also included.
It was not within the scope of this work to develop new training data sets. However, in identifying tools that have not been tested in a natural history context, we do highlight where the construction and application of new training datasets needs to be prioritised. Ground truth datasets will become increasingly important for natural history data, especially as the variety and specificity of segmentation become more complex.
1.1.2 Prior Research on Automation
A collection of research has previously been conducted in the EC funded SYNTHESYS3 and ICEDIG projects on the capabilities of automation tools in digitisation. For the former,
1.1.3 Crowdsourcing and Human-in-the-Loop
As the Specimen Data Refinery is intended to integrate both artificial intelligence (AI) and human-in-the-loop (HitL) approaches to extraction and annotation, citizen science platforms such as plant identification apps and volunteer transcription services were included in the initial research. However, the primary focus of this landscape analysis is on AI platforms as these hold the greatest untapped potential for mass efficiency gains and centralised workflows.
1.2 Project Context
This report was adapted from a formal Deliverable (D8.1) of the SYNTHESYS+ project that was previously made available to project partners and submitted to the European Commission as an internal report. While the differences between these versions are minor the authors consider this the definitive version of the report.
This paper is a precursor to the development of new tools, services, workflows and a formal registry, which form the basis of the next SDR task (8.2) in the SYNTHESYS+ project. We hope this report will be broadly useful for software development teams and informatics teams outside of the SYNTHESYS+ project working on new software-based approaches to improve mass digitisation of natural history specimens.
2. Methodology
In order to collect an aggregated list of tools to evaluate, the SYNTHESYS+ partners from partner institutions were invited to contribute known tools, methods, resources and pilot projects (Suppl. material
- Brief service description
- Delivery platform (eg. web application, software library, R package, etc.)
- Associated academic papers
- Known test pilots
- Cost (where applicable)
- Input/Output formats
- License
In total, 76 tools, methods and resources were collected.
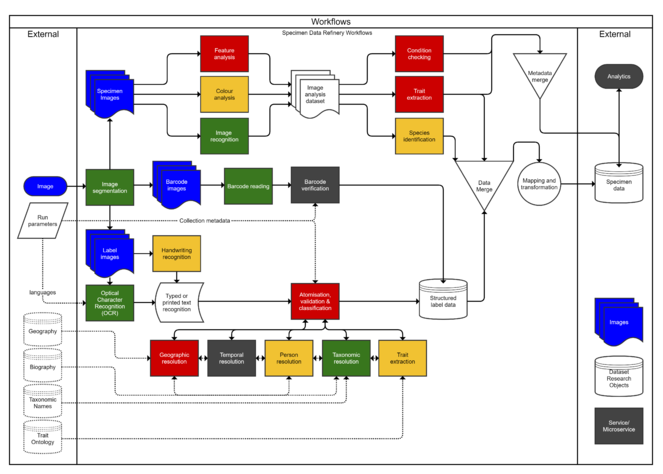
After the aggregation phase was complete, the list was reviewed in its entirety. Each tool and resource was mapped onto the data refinery workflow, in order to assess where reusable resources are available, and where there are major gaps or potential risks. Each step in the workflow was graded according to a traffic-light system - green for the existence of a variety of resources that could be repurposed, amber for the existence of resources with limited reuse potential, and red for a major gap where either no resource exists or there is no reuse potential. A number of steps in the workflow (identifier verification, trait extraction and analytics) had no associated tools submitted and were marked as grey in the workflow map. The workflow map was then distributed to the contributing partners to identify any further gaps or missing areas.
Upon completion of the gap analysis, an initial assessment was conducted on the technology stack available to compile each of the tools together into a workflow. A high-level consultation was conducted with a computer science team at a partner institution with prior experience developing similar complex human-in-the-loop workflows. Their recommendations have been documented for further study and research in the next phase.
3. Gap Analysis
This analysis revealed that there are some areas where considerable efforts have been put towards developing a toolkit, while others have received less efforts (see Fig.
Traffic-light (RAG) analysis of tools and services: green = existence of a variety of resources that could be repurposed; amber = existence of resources with limited reuse potential; red = a major gap where either no resource exists or there is no reuse potential.
| Tool/Service Functionality | Traffic-light Status |
|---|---|
| Image segmentation | Green |
| Feature analysis | Red |
| Colour analysis | Amber |
| Image recognition (object detection) | Green |
| Condition checking | Red |
| Trait extraction | Red |
| Species identification | Amber |
| Handwritten text (handwriting) recognition | Amber |
| OCR of typed or printed text | Green |
| Atomisation, validation and classification | Red |
| Geographic resolution | Red |
| Person resolution | Amber |
| Taxonomic resolution | Green |
| Label (biological) trait extraction | Amber |
3.1 Image segmentation
Image segmentation involves dividing the pixels of an image into its component parts, such as separating the specimen itself from the barcode and the label. Image segmentation is a fundamental low-level image processing task, which faciliates subsequent higher-level tasks on the resulting components, such as image object detection and recognition. In addition to a large suite of tools available for batch photo editing (cropping, resizing, rotating, etc.), there were three reported tools that could segment an image. scikit-image (
There is some overlap between semantic segmentation and object detection (see Section 3.2). Semantic segmentation refers to a type of segmentation where each pixel is labelled as belonging to a particular class of objects. It is similar to image object detection and recognition and goes beyond the individually labelled pixel level to recognise entire objects in an area. While YOLO V3 is an object detection tool, it has been used to identify and segment the different objects that are commonly found on herbarium sheets: the pressed plant specimen, scale bar, stamp, color calibration chart, specimen label, envelope and barcode (
The step was marked as green because there is more than one tool available and each tool provides a different method for going about image segmentation, thus offering a variety of options that could be tested based on the needs of the collection’s images. More importantly, scikit-image underwent significant testing by the Natural History Museum as part of the SYNTHESYS3 project (
3.2 Feature analysis, colour analysis and image recognition (object detection)
In the aggregation process, many tools were listed as feature analysis resources (e.g. Flavia and LeafProcessor) but were ultimately categorised as species identification tools because they used some level of feature analysis to identify a specimen (primarily plants). The only tool used for broader feature analysis in natural history specimens was a set of prototypes developed in the SYNTHESYS3 project which segments the specimen from the background of the image by identifying its edges and then, for butterflies and moths only, takes measurements of the wings (
Colour analysis was categorised as amber because there is only one tool available. Image Quality Assessment, in addition to predicting the technical quality of the image, is able to group sets of images together based on similar colours. However, other tools used for image segmentation and recognition, like scikit-image, may be used for this.
Image recognition was categorised as green because there are two well-developed and heavily-supported resources available. Google Vision comes with enterprise-level support and longevity and offers toolkits for both non-coders and programmers. OpenCV has a strong open-source development infrastructure underneath it. Both can be trained to recognise items in an image and organise them into pre-set categories.
3.3 Condition checking, image trait extraction and species identification
No tools or resources were submitted for condition checking and this appears to be a major gap in the workflow. In the context of the SDR this would be a series of varying visual checks on a natural history specimen that may cover their stability, damage, completeness and potential for use.
A majority of the image trait extraction tools and resources developed have been for biomedical/epidemiological purposes. Trait extraction was marked as red because only two tools were submitted and both are applicable only to plants. Plant Trait Extraction is capable of phenotypic trait extraction but only for a subset of collections (
Species identification, in contrast, has received a tremendous amount of concerted effort, research and R&D. As a result, numerous tools and methods have been developed spanning the range of neural network machine learning tools (
3.4 Optical character recognition of handwritten and printed/typed text
These areas have also been the recipients of considerable research and development. While Transkribus is the only listed tool available for handwritten text transcription and analysis, it is supported by EU funding and has been successfully deployed on a collection of specimens from the Royal Botanic Garden Edinburgh. Transkribus also offers a host of web and cloud services.
OCR in general was marked green as there are multiple tools available, although ABBYY is an enterprise-level software that will likely have cost associated. The Natural History Museum, London has tested Tesseract 4.1.0 OCR (
While yet to be tested, certain handwriting such as the signatures of prolific collectors may be more reliably identified using image recognition rather than OCR. There may also be other repetitive words or phrases, such as name prefixes and stamps indicating nomenclatural type status, which could be identified in this way.
3.5 Atomization, validation and classification
Many OCR tools are capable of named entity recognition (NER) - the ability to extract strings of text and thereby break a label into its component parts, such as place names, person names or taxon names. The main tools - NLTK, spaCy, flair, and Stanford Named Entity Recognizer - are capable of deep learning so can be trained to recognise specific strings and categories from a ground truth dataset. These tools have been used to derive structured data from taxonomic publications (e.g. traits), but still require further research in the context of natural history collection labels. There are also a couple of tools available for extracting ecologically-relevant terms from a label. ClearEarth and Explorer of Taxon Concepts are both capable of identifying such terms and categorising (
There are also a number of language detection tools available (
3.6 Geographic resolution, person resolution and taxonomic resolution
Geographic resolution is a task natural history collections have struggled to automate. There are numerous tools available for general geocoding - MapQuest Geocoding, Google Geocoding, CartoDB, Pelias. However these tools require a known address, city, country or region name in order to identify an associated latitude and longitude. They are not designed for historical place names and cannot accommodate changing boundaries over time or vague or general place descriptions. GEOLocate is the only tool listed that is designed specifically to assist in the geographic resolution of natural history collections and is currently still active. A number of other tools like BioGeomancer (
Person resolution was marked as amber because Bionomia (formerly Bloodhound) is currently the only tool designed specifically to match a collector with the specimens they collected. Numerous efforts are also underway to assign unique person identifiers to researchers, present-day and historical. ORCID, ISNI and ResearcherID have databases of person identification numbers and VIAF combines the person with numerous countries’ national libraries into an aggregated database. In relation to published academic papers, Elsevier assigns a researcher ID for all authors in its database through Scopus and there are a number of sites to which researchers can upload their publishing profile.
The Muséum national d'Histoire naturelle is currently developing a Person Refinery, expected to be completed by April 2020, which has revealed a number of challenges in efficiently developing data structures and alignments for person resolution, chief of which is how the various researcher ID systems can help disambiguate person names within collections and whether there is particular people identifier system which will prove to be most relevant for all types of collections (
Taxonomic name resolution is the most developed. There are many tools available, and there is an increasing level of integration of tooling into various initiatives. The Catalogue of Life is an authoritative global species checklist for all life on earth, that is built on 172 global taxonomic resources (
Taxonomic name resolution is marked green as there are a number of tools and resources available, and also the level of integration is relatively well developed. However there are still several challenges. Taxonomic gaps exist, taxonomic data sources may portray alternative classifications, and it is not always clear if datasources have been build on the same nomenclatural foundation. This may result in a scattered and blurry landscape for users, but at the same time highlights the importance of both scientific names and taxa (
3.7 Label (Biological) Trait Extraction
Biological trait extraction has been largely confined to text mining of literature (
4. Building a Workflow
The Specimen Data Refinery (SDR) aims to take a selection of tools identified above and package them into a cohesive workflow for processing and analysis. This requires a technology stack that will create the links between different tools and the operating environment in which the workflow is executed and managed. While there are many different technology services available for workflow development, the priority for the SDR will be to identify a technology stack that contains all of the required functionality, while being reliable, sustainable and cost-effective.
4.1 Selecting a Human-in-the-Loop Workflow Management Systems
There are many examples in bioinformatics of automated workflows that string together a collection of tools and execute a series of steps with no intervention required by a user (
There are currently over 280 Workflow Management Systems, each with their own strengths and weaknesses. Typically, they vary on whether they are focused on linking tools or linking infrastructure layers; whether they are domain-specific or general; and who they target as their user-base and the level of expertise required. It is not necessary, however, to choose only one. Workflow management systems can be combined to develop custom solutions.
In addition to these considerations, the SDR has an added layer of complexity not represented in Fig.
Therefore, the environment within which the workflow is executed should support interactivity, providing a space in which a user can give commands that then dictate the next steps of the workflow. Similar HitL workflows have been developed for other biodiversity projects (
Galaxy is another WfMS designed specifically for bioinformatics that offers HitL functionality (
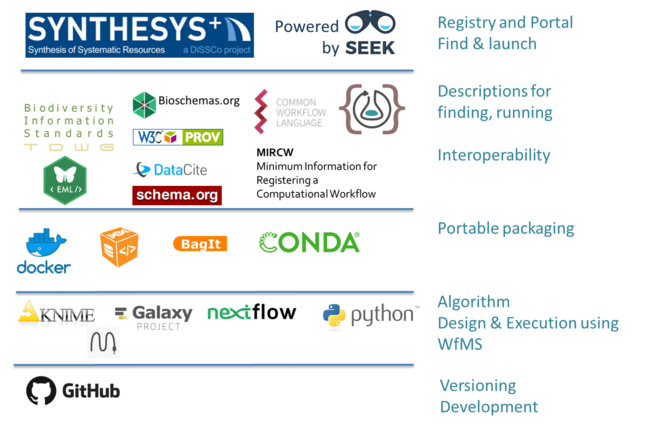
4.2 Implementing a standardised workflow language for interoperability
The steps of a workflow (scripts, tools, command-line tools and workflows themselves) are linked together and executed by the workflow engine within the WfMS. Linking all of these disparate interfaces, scripts, methods and datasets together requires each step to be in the same workflow language with interoperable data standards so that they can communicate consistently with each other.
Different WfMS typically have different language requirements and protocols, and limit interoperability. Several attempts have been made to standardise workflow descriptions and enable workflow interoperability between different systems in order to support the long-term preservation of workflows that may outlive any specific WfMS. The Workflow Description Language and the Common Workflow Language (CWL) (
The Common Workflow Language is an open standard for compiling workflows and describing how to run the command line tools inside them in a way that makes them portable and scalable. It is a WfMS-agnostic common language that developers can use to better document workflows and assist with workflow portability and interoperability when working between different systems. The current CWL Standard (v1.1) provides authoritative documentation of the execution of CWL documents.
ELIXIR, a sister ESFRI to DiSSCo, has invested in the support of CWL and it is used by the EU’s BioExcel2 Centre of Excellence for Biomolecular modelling, by the IBISBA ESFRI for Industrial Biotechnology and by the EOSCLife cluster project. This strong community and financial support for the development of CWL is indicative of its longevity and anticipated sustainability which are important factors when deciding which workflow language to use in the SDR.
4.3 Incorporating prior information and the statistical framework
The SDR should not work independently from prior knowledge. Some basic information on the collection is always known about specimen images before they are fed into the SDR. These data are generally known as collection metadata. These data might be the taxonomic scope of the collection (e.g. insects, vascular plants, fungi), geographic scope (e.g. Belgium, Asia, Berkshire), date range, etc. Often folders and boxes of images are created together and additional metadata from these batches are captured during imaging. All of these data provide prior information that, with the right statistical framework, can considerably improve the outcome of the SDR. These data can be used as informative priors in a decision tree to direct the images to a suitably trained AI system. However, they might also be used after AI processing either to validate the output or by combining the probabilities from independent processes. There is also prior information about the nature of images to be processed, such as the camera and lighting used, the approximate size of the object and the orientation of the object to the camera (
Even in the absence of any prior knowledge of the origin and identification of the specimen, the who, what, when and where of a specimen are all interconnected. Biographies tell us what, when and where collectors are likely to have collected; and known species distributions tell us what countries they are likely to be from. So, where the country is determined from the label with 90% accuracy, for instance, this information could be used in further processing to make the determination of the collector, date and taxon more reliable.
Prior knowledge needs to be combined with derived information to generate the final result. For example, imagine a European butterfly collection digitized by imaging both its label and a dorsal view of the insect. The image of the insect is processed through an AI to determine its identity and the label is processed through OCR, followed by entity recognition to find a taxonomic name. How are these two determinations of the taxonomic identity of the specimen combined into a reliable output? Also, how can we use the prior knowledge that this was a European butterfly collection to improve the AI, OCR and entity recognition output while making it transparent to an end user about how such identifications were made
It remains a considerable challenge to create a workflow that incorporates prior knowledge, uses learned knowledge, propagates uncertainty in the workflow and outputs the result with a value for certainty. While, so far, such workflows have not been currently addressed in biodiversity science, there has been research in this area in large-scale microscopic analysis used for diagnoses (
4.4 Assembling the workflow
Workflows are made up of a collection of metadata and files - test data, example data, validation data, design documents, parameter files, parameter setting files, result files, provenance logs, etc (
While the Common Workflow Language is the language in which a workflow is written and described, a research object (RO) is a method for packaging and linking the metadata of disparate scholarly information using certain standards and conventions so that the packages can be exported and exchanged between WfMSs with the necessary detail to be reused and reproduced (
Along with the CWL and Galaxy, RO-Crate has been adopted by EOSCLife and IBISBA as the service for describing and packaging workflows and their related files. Based on this community and financial support for these capabilities, a number of WfMSs, including Galaxy, will support CWL and RO-Crate.
4.5 The Specimen Data Refinery techology stack
Executing the SDR workflow will require a foundational tech stack and infrastructure for two core pieces - a registry and a run platform.
A registry is a library of workflows. All of the tools and steps in the workflow will be comprised of smaller sub-steps and sub-workflows that make up the building blocks of the entire engine. These building blocks will be housed in a registry built for the SDR. WorkflowHub is a workflow library currently under development for EOSCLife and IBISBAHub for IBISBA workflows. SEEK, the underlying platform for both of these Hubs, can also be utilized for the SDR (
A run platform is the technology stack that will pull all of these tools, services and processes together. Along with a variety of other services, the recommended SDR run platform (Fig.
5. Conclusion
This gap analysis has made apparent which categories of tools and resources have been specifically developed for specimen images or can be readily generalised and potentially used. Image segmentation, OCR and taxonomic resolution have a broad range of existing and well-tested approaches. Other areas such as visual trait extraction or text processing tools to convert “strings to things” are lacking. There are some general tools and commercial services which deal with contemporary languages but Latin and Greek are commonly encountered in scientific names, in diagnostic descriptions (especially botanical descriptions) and as abbreviations on labels such as “cf.” (confer). Other potential issues that are yet to be tested or understood are the frequency of co-occurring languages on labels; the frequency of differing co-occurring handwriting (also known as "hands") on labels; and how challenging the abbreviated technical writing style of labels is compared to natural language documents.
Many of the tools and services will require initial or further testing and analysis with training datasets that are domain-specific to natural history collections, in order to assess their quality and accuracy. For example, the Botanic Garden Meise recently undertook an image recognition pilot with Google Vision to extract label information but the results have yet to be analysed for accuracy (
While there are a broad selection of taxonomic name resolution tools and services, many of which are incorporated into GBIF’s name backbone (
We expect to develop training datasets for the following components of the SDR workflow:
- Image segmentation
- Image recognition
- Feature analysis
- Trait extraction
- Condition checking
- Species identification
- Atomisation, validation and classification
- Person and geographic resolution
However, the development of ground-truth training data sets requires considerable time and resources (
Previous projects to develop toolsets or platforms, like BioGeomancer, have suffered from sustainability issues after project funding ceased. We may find that some tools or scripts have performance issues in the SDR if used at scale. Tools and datasets developed in the next phase of SDR work should prioritise software sustainability. Considerations for sustainability include making use of existing standards, comprehensive functional and high-throughput performance/scaling tests, service/tool documentation, and having a maintenance plan - these are summarised in detail by the Software Sustainability Institute. In terms of workflow platform sustainability, we should use a pre-vetted platform, ideally with hosting support, that makes use of existing European investment and prior efforts in training, notably in the ESFRI Cluster EOSCLife and the ESFRI IBISBA.
The efficiency of the SDR will come from large-scale processing of images and specimen data. Images, particularly high resolution and lossless formats, are large files. Transferring, retrieving, sharing and storing the originals and their derivatives is likely to be slow and potentially expensive. This is one of the most important issues that the SDR will need to address, with careful consideration of downsampling, subsampling, overall file size and the number of transfers. While a cloud-based solution is desirable we are likely to need to offer locally hosted solutions to avoid prohibitive costs.
While all of these complexities and hurdles need to be taken into consideration in developing the SDR, this analysis also revealed there is a considerable amount of software already available, both open source and proprietary, and research that has already been conducted into automating many of these processes. There is significant opportunity to take advantage of this research by combining it into a workflow that will greatly improve the efficiency and scalability of natural history digitisation efforts.
Glossary
Active contouring: a method of image segmentation that identifies object contours in an image in order to detect outlines.
Condition checking: a series of varying checks on a natural history specimen that may cover their stability, damage, completeness and potential for use. Some examples include: visually checking mountant colour in microscope slides to determine mountant type and need for remounting, presence and severity of verdigris in entomological specimens or pyrite decay in paleontological specimens.
ETL: extract, transform, load - usually used to describe the process of extracing data from one (or more) database/system then transforming it so it can be loaded it into another.
GBIF: Global Biodiversity Information Facility (https://www.gbif.org/).
Google Vision: a machine learning tool for automated image recognition and categorisation (https://cloud.google.com/vision).
Ground truth data: a dataset comprised of information acquired through direct observation rather than through inference or automation.
Hands: handwritten script attributable to an individual/individuals.
ICEDIG: EC-funded project "Innovation and consolidation for large scale digitisation of natural heritage" (https://www.icedig.eu).
Image recognition: software to identify the contents of an image, including objects, locations, text and actions being performed.
Metadata: a set of data that describes and gives information about other data, such as the file format of timestamp of an image or the provenance and processing inputs of a data run.
Neural network: a set of algorithms that are designed to recognize patterns and connections through training on a dataset (see training dataset).
NLP: natural language processing - software to understand human natural language including contextualisation and semantics.
OCR: optical character recognition - software to convert images of typed or handwritten text into machine readable encoded text.
Reference datasets: data that sets standards to which the fields in other datasets adhere.
RO: research object - a rich aggregation of resources used in a scientific investigation and/or to provide comprehensive supporting information for a published paper with the aim to improve reproducibility (http://www.researchobject.org).
SDR: Specimen Data Refinery.
SEEK: a digital object management and cataloguing platform that underpins the Workflow Hub and IBISBAHub.
Thresholding: a method for segmenting an image by converting a colour image to grayscale and then filtering out pixels that are above a certain setting on the grayscale - a threshold - and maintaining pixels that fall below it.
Training datasets: datasets that are used to train a machine learning platform in a particular set of capabilities, for example to identify something in an image.
Trait extraction: automated processes to identify and quantify specific characteristics of an organism, most likely phenotypic data.
WfMS: workflow management system.
YOLO V3: the third release of “You only look once”, an tool for detecting images in an object and segmenting them.
Acknowledgements
LL would like to thank: James Durrant for discussing and developing the initial concept and early prototypes of Specimen Data Refinery services; Matt Woodburn for creating the initial diagram and discussions on service implementation; comments and proof reading by the report contributors and by Helen Hardy; review feedback from Mark Hereld and Rebecca Dikow.
Funding program
Grant title
SYNTHESYS+ (submitted as SYNTHESYS PLUS), Grant agreement ID: 823827
Author contributions
Authors:
Stephanie Walton: Data Curation, Investigation, Methodology, Visualization, Writing – Original Draft. Laurence Livermore: Conceptualization, Data Curation, Investigation, Visualization, Supervision, Writing – Original Draft, Writing – Review & Editing. Olaf Banki: Data Curation, Investigation, Writing – Review & Editing. Robert W. N. Cubey: Data Curation, Investigation, Writing – Review & Editing. Robyn Drinkwater: Data Curation, Investigation, Writing – Review & Editing. Markus Englund: Investigation, Writing – Review & Editing. Carole Goble: Conceptualization, Investigation, Visualization, Writing – Original Draft. Quentin Groom: Resources, Investigation, Visualization, Writing – Original Draft, Writing – Review & Editing. Christopher Kermovant: Investigation, Writing – Review & Editing. Isabel Rey: Investigation, Writing – Review & Editing. Celia M Santos: Investigation, Writing – Review & Editing. Ben Scott: Investigation, Writing – Review & Editing. Alan R. Williams: Conceptualization, Investigation, Writing – Original Draft. Zhengzhe Wu: Investigation, Writing – Review & Editing.
Contributors:
The following people contributed (Investigation) to the tools and services dataset and/or made suggestions on papers/software for background research: Mathias Dillen, Elspeth Haston, Matthias Obst, Mario Lasseck, Nicky Nicholson, Sarah Phillips, Dominik Röpert.
Contribution types are drawn from CRediT - Contributor Roles Taxonomy.
References
-
The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update.Nucleic Acids Research46https://doi.org/10.1093/nar/gky379
-
Common Workflow Language.1.0.Common Workflow Language working group. URL: http://doi.org/10.6084/m9.figshare.3115156.v2
-
Approaches to estimating the universe of natural history collections data.Biodiversity Informatics7(2). https://doi.org/10.17161/bi.v7i2.3991
-
Catalogue of Life Plus: A collaborative project to complete the checklist of the world's species.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37652
-
Using a suite of ontologies for preserving workflow-centric research objects.Journal of Web Semantics32:16‑42. https://doi.org/10.1016/j.websem.2015.01.003
-
People of Collections: Facilitators of Interoperability?Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.35268
-
taxize: taxonomic search and retrieval in R.F1000Research2https://doi.org/10.12688/f1000research.2-191.v2
-
Scientific workflows for computational reproducibility in the life sciences: Status, challenges and opportunities.Future Generation Computer Systems75:284‑298. https://doi.org/10.1016/j.future.2017.01.012
-
Introducing Explorer of Taxon Concepts with a case study on spider measurement matrix building.BMC Bioinformatics17(1). https://doi.org/10.1186/s12859-016-1352-7
-
Enabling machine-actionable semantics for comparative analyses of trait evolution.Zenodohttps://doi.org/10.5281/zenodo.885538
-
langdetect. URL: https://pypi.org/project/langdetect
-
The future of scientific workflows.The International Journal of High Performance Computing Applications32(1):159‑175. https://doi.org/10.1177/1094342017704893
-
A benchmark dataset of herbarium specimen images with label data.Biodiversity Data Journal7https://doi.org/10.3897/bdj.7.e31817
-
Computer Vision for biological specimen images.Natural History Museum. URL: https://github.com/NaturalHistoryMuseum/vision
-
Extraction of phenotypic traits from taxonomic descriptions for the tree of life using natural language processing.Applications in Plant Sciences6(3). https://doi.org/10.1002/aps3.1035
-
Long-term preservation of herbarium specimen images. https://www.eudat.eu/communities/herbadrop. Accessed on: 2020-5-11.
-
Measuring Morphological Functional Leaf Traits From Digitized Herbarium Specimens Using TraitEx Software.Biodiversity Information Science and Standards3https://doi.org/10.3897/biss.3.37091
-
GBIF Occurrence Download. https://doi.org/10.15468/dl.8pq57z. Accessed on: 2020-6-08.
-
GBIF Backbone Taxonomy. Checklist dataset.GBIF. URL: https://doi.org/10.15468/39omei
-
FAIR Computational Workflows.Data Intelligence2:108‑121. https://doi.org/10.1162/dint_a_00033
-
BioGeomancer: Automated Georeferencing to Map the World's Biodiversity Data.PLoS Biology4(11). https://doi.org/10.1371/journal.pbio.0040381
-
D4.2 - Automating data capture from natural history specimens. http://synthesys3.myspecies.info/node/695. Accessed on: 2020-5-11.
-
iNaturalist Computer Vision Explorations. https://www.inaturalist.org/pages/computer_vision_demo. Accessed on: 2020-6-16.
-
Automatic Plant Identification: Is Shape the Key Feature?Procedia Computer Science76:436‑442. https://doi.org/10.1016/j.procs.2015.12.287
-
ecocore.20200518. Release date:2020-5-18. URL: https://github.com/EcologicalSemantics/ecocore
-
Stem–Leaf Segmentation and Phenotypic Trait Extraction of Individual Maize Using Terrestrial LiDAR Data.IEEE Transactions on Geoscience and Remote Sensing57(3):1336‑1346. https://doi.org/10.1109/tgrs.2018.2866056
-
Sharing interoperable workflow provenance: A review of best practices and their practical application in CWLProv.GigaScience8(11). https://doi.org/10.1093/gigascience/giz095
-
Image-based Plant Species Identification with Deep Convolutional Neural Networks. In:CLEF 2017 Working Notes,1886.Conference and Labs of the Evaluation FOrum,Dublin, Ireland,11-14 September 2017.CLEF[InEnglish]. URL: http://ceur-ws.org/Vol-1866/
-
langid.py: An Off-the-shelf Language Identification Tool. In:Proceedings of the ACL 2012 System Demonstrations. ,Jeju Island, Korea,2012.Association for Computational LinguisticsURL: https://www.aclweb.org/anthology/P12-3005/
-
A semi-automated workflow for biodiversity data retrieval, cleaning, and quality control.Biodiversity Data Journal2https://doi.org/10.3897/bdj.2.e4221
-
Discovery and publishing of primary biodiversity data associated with multimedia resources: The Audubon Core strategies and approaches.Biodiversity Informatics8(2). https://doi.org/10.17161/bi.v8i2.4117
-
Plant Leaf Recognition Using Shape Features and Colour Histogram with K-nearest Neighbour Classifiers.Procedia Computer Science58:740‑747. https://doi.org/10.1016/j.procs.2015.08.095
-
Leaf recognition of woody species in Central Europe.Biosystems Engineering115(4):444‑452. https://doi.org/10.1016/j.biosystemseng.2013.04.007
-
Methods for Automated Text Digitisation. https://doi.org/10.5281/zenodo.3364502. Accessed on: 2020-2-27.
-
Towards a scientific workflow featuring Natural Language Processing for the digitisation of natural history collections.Research Ideas and Outcomes6https://doi.org/10.3897/rio.6.e55789
-
FreeLing 3.0: Towards Wider Multilinguality. In:Proceedings of the Eighth International Conference on Language Resources and Evaluation (LREC'12).Eighth International Conference on Language Resources and Evaluation (LREC'12),Istanbul, Turkey,2012.European Language Resources Association (ELRA)URL: http://www.lrec-conf.org/proceedings/lrec2012/pdf/430_Paper.pdf
-
Image Segmentation using Python’s scikit-image module. https://towardsdatascience.com/image-segmentation-using-pythons-scikit-image-module-533a61ecc980. Accessed on: 2020-2-26.
-
Challenges with using names to link digital biodiversity information.Biodiversity data journal4:e8080. https://doi.org/10.3897/BDJ.4.e8080
-
Machine Learning Using Digitized Herbarium Specimens to Advance Phenological Research.BioSciencehttps://doi.org/10.1093/biosci/biaa044
-
Workflow systems turn raw data into scientific knowledge.Nature573(7772):149‑150. https://doi.org/10.1038/d41586-019-02619-z
-
The use and limits of scientific names in biological informatics.ZooKeys550:207‑223. https://doi.org/10.3897/zookeys.550.9546
-
biogeo: Point Data Quality Assessment and Coordinate Conversion.1.0.CRAN. Release date:2016-4-08. URL: https://cran.r-project.org/package=biogeo
-
Biogeo: an R package for assessing and improving data quality of occurrence record datasets.Ecography39(4):394‑401. https://doi.org/10.1111/ecog.02118
-
U-Net: Convolutional Networks for Biomedical Image Segmentation.Lecture Notes in Computer Science234‑241. https://doi.org/10.1007/978-3-319-24574-4_28
-
Species 2000 & ITIS Catalogue of Life, 2019 Annual Checklist. http://www.catalogueoflife.org/annual-checklist/2019/. Accessed on: 2020-5-11.
-
Interface to the "Geonames'' Spatial Query Web Service.0.999.CRAN. Release date:2019-2-19. URL: https://github.com/ropensci/geonames
-
Leaves Recognition System Using a Neural Network.Procedia Computer Science102:578‑582. https://doi.org/10.1016/j.procs.2016.09.445
-
SYNTHESYS+ Abridged Grant Proposal.Research Ideas and Outcomes5https://doi.org/10.3897/rio.5.e46404
-
New Methods to Improve Large-Scale Microscopy Image Analysis with Prior Knowledge and Uncertainty.KIT Scientific Publishing,Karlsruhe,243pp. [ISBN978-3-7315-0590-7] https://doi.org/10.5445/KSP/1000060221
-
Fuzzy-based propagation of prior knowledge to improve large-scale image analysis pipelines.PLOS ONE12(11). https://doi.org/10.1371/journal.pone.0187535
-
Tesseract OCR.4.1.0.GitHub. Release date:2019-7-07. URL: https://github.com/tesseract-ocr/tesseract/releases/tag/4.1.0
-
Automated Trait Extraction using ClearEarth, a Natural Language Processing System for Text Mining in Natural Sciences.Biodiversity Information Science and Standards2https://doi.org/10.3897/biss.2.26080
-
Objects Detection from Digitized Herbarium Specimen based on Improved YOLO V3.Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applicationshttps://doi.org/10.5220/0009170005230529
-
GBIF Name Resolution.Natural History Museum. Release date:2020-6-17. URL: https://github.com/NaturalHistoryMuseum/gbif-name-resolution
-
A cost analysis of transcription systems.In prep..
-
Generating segmentation masks of herbarium specimens and a data set for training segmentation models using deep learning.Applications in Plant Sciences8(6). https://doi.org/10.1002/aps3.11352
-
SEEK: a systems biology data and model management platform.BMC Systems Biology9(1). https://doi.org/10.1186/s12918-015-0174-y
-
A Leaf Recognition Algorithm for Plant Classification Using Probabilistic Neural Network.2007 IEEE International Symposium on Signal Processing and Information Technologyhttps://doi.org/10.1109/isspit.2007.4458016
-
The Effect of Background on A Deep Learning Model in Identifying Images of Butterfly Species.Electrical and Electronics Engineering: An International Journal8(1):01‑08. https://doi.org/10.14810/elelij.2019.8101
Supplementary material
Evaluation of 89 tools and services with a categorisation, summary, cost information, pilot data, software status, input format(s), output format(s), comments and license.
Between 6.2% and 12.5% of specimens are digitised and publicly available on GBIF based on the total number of estimated natural history specimens by