|
Research Ideas and Outcomes :
Research Idea
|
|
Corresponding author: Sheeba Samuel (sheeba.samuel@uni-jena.de), Maha Shadaydeh (maha.shadaydeh@uni-jena.de)
Received: 09 May 2020 | Published: 11 May 2020
© 2020 Sheeba Samuel, Maha Shadaydeh, Sebastian Böcker, Bernd Brügmann, Solveig Franziska Bucher, Volker Deckert, Joachim Denzler, Peter Dittrich, Ferdinand von Eggeling, Daniel Güllmar, Orlando Guntinas-Lichius, Birgitta König-Ries, Frank Löffler, Lutz Maicher, Manja Marz, Mirco Migliavacca, Jürgen R. Reichenbach, Markus Reichstein, Christine Römermann, Andrea Wittig
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Samuel S, Shadaydeh M, Böcker S, Brügmann B, Bucher SF, Deckert V, Denzler J, Dittrich P, von Eggeling F, Güllmar D, Guntinas-Lichius O, König-Ries B, Löffler F, Maicher L, Marz M, Migliavacca M, R. Reichenbach J, Reichstein M, Römermann C, Wittig A (2020) A virtual “Werkstatt” for digitization in the sciences. Research Ideas and Outcomes 6: e54106. https://doi.org/10.3897/rio.6.e54106
|
|
Abstract
Data is central in almost all scientific disciplines nowadays. Furthermore, intelligent systems have developed rapidly in recent years, so that in many disciplines the expectation is emerging that with the help of intelligent systems, significant challenges can be overcome and science can be done in completely new ways. In order for this to succeed, however, first, fundamental research in computer science is still required, and, second, generic tools must be developed on which specialized solutions can be built. In this paper, we introduce a recently started collaborative project funded by the Carl Zeiss Foundation, a virtual manufactory for digitization in the sciences, the “Werkstatt”, which is being established at the Michael Stifel Center Jena (MSCJ) for data-driven and simulation science to address fundamental questions in computer science and applications. The Werkstatt focuses on three key areas, which include generic tools for machine learning, knowledge generation using machine learning processes, and semantic methods for the data life cycle, as well as the application of these topics in different disciplines. Core and pilot projects address the key aspects of the topics and form the basis for sustainable work in the Werkstatt.
Keywords
Machine learning, semantic methods, data management
Overview and background
Data plays an increasingly important role in many scientific disciplines. They are produced almost everywhere, but their use is still limited. This is mostly due to the fact that in many cases manual analysis is no longer possible due to the large data volume, and automatic analysis is not yet possible due to the lack of tools and/or expertise by the scientists. Here, intelligent systems combined with machine learning promise a solution on how to extract knowledge and how to carry out different steps in the process of the data life cycle from data generation and analysis to storage and subsequent use. This can be done either fully automatically or in collaboration between human and machine. For this vision to become reality, essential challenges need to be tackled. In this paper, we provide an overview of a recently launched project, the digitization “Werkstatt*
Objectives
The “Werkstatt” addresses the following three main research topics in computer science (T1-T3) as well as the application fields (T4):
T1. Generic tools for machine learning
An increasingly important topic within machine learning is its automation (AutoML). First steps are the preparation of the data, e.g., by transformation into suitable data formats, the treatment of missing values, or the correction of erroneous data entries. Moreover, most algorithms are highly parameterized and require a good choice of parameter values, e.g., the architecture of a deep neural network. The algorithm must be executed and its results analyzed, with the question of choosing appropriate metrics and measures of success. The analysis may require additional data to be collected later. These diverse, manual optimization steps usually require methodological expertise. Therefore, one goal is to adopt novel methods of AutoML design by using a systematic search. Another challenge for the successful use of machine learning is when the question to be answered by the data is not fixed a priori. One way to deal with this is to learn a flexible, generic model that a human analyst can examine with a suitable visual interface. For this purpose, due to their generality, multivariate probabilistic models are very suitable. A third important area for the development of generic tools for machine learning is the provision of automatic differentiation methods. Frequently, derivatives of an objective function to be optimized are required during learning. Automatic differentiation enables the automated generation of computer programs that calculate first or higher order derivatives of such target functions. However, typical target functions in machine learning have special structures that are not used in standard automatic differentiation methods. A goal that should, therefore, be pursued in this area is, in general, techniques of automatically distinguishing specific structures from the machine to adapt to learning. Such targeted program transformation could result in programs that require significantly less space or whose execution time is significantly shorter than general automatic differentiation techniques.
T2. Knowledge generation using machine learning processes
Machine learning methods, in particular supervised learning methods, have been able to learn input/output relationships from data for years and thus solve problems such as classification or regression tasks with high accuracy. In the field of image and scene analysis, the achieved accuracy reaches or surpasses the performance of humans (
With all the advances in generating the correct input/output relationship, challenging issues still exist in data-driven solutions. Scientists who want to generate knowledge from data, would like to make a model refinement via data-model integration and if necessary, switch between hypothesis- and data-driven procedures. Despite the tremendous power of deep neural networks, the following questions are still open and need extensive research efforts: a) Can the machine learning process be understood and reconstructed? b) Is it possible to detect cause-effect relationships by data-driven methods and thus to identify causal relationships for example in multivariate time series? c) How to integrate knowledge about, e.g., physical laws, with a data-driven approach to improve the approximation accuracy of the data estimated model and at the same time reduce the effort required for training (data collection, data annotation, the number of learning steps, etc.)? d) Can a large amount of annotated training data, which are still required for many approaches, be reduced in order to open up new fields of applications, in which such data collection is currently too expensive or completely impossible? In this Werkstatt, we aim to add new contributions to the line of tackling these issues. The main focus will be on the development of unsupervised learning methods as well as data-driven solutions for causal inference in dynamical systems.
T3. Semantic methods for the data life cycle
The added value of digitizing the research processes arises not only when the individual steps are improved by intelligent systems, but also when integrated support exists throughout the entire scientific (or data) life cycle. Semantic methods offer promising approaches here. An essential component of digitization of research processes is a precise and machine-understandable description of the experimental data, which helps in the retrieval and re-use of data, the reproducibility of the results, and more generally, scientific progress. However, scientists are reluctant to provide the descriptions because this requires high effort and time. One goal of this topic is to investigate how experiment data can be semi-automatically annotated with semantic metadata with the support of artificial intelligence. Another aspect is the role of data curation and data quality control in this environment. The quality of the research results depends heavily on the quality of the data. However, in the face of a rapidly growing amount of data, the major dilemma is that a high curation level for all the data is not realistic and not cost-effective. In this Werkstatt, we aim to develop novel methods for determining the data value and integrate the methods for the control of curation processes into systems. The main focus will be on the construction and maintenance of knowledge graphs.
T4. Applications
The methods developed in T1-T3 are intended to be used, evaluated and optimized directly in fields of application and enable discipline-specific breakthroughs. The development of fundamentals in machine learning and data management in the sciences can only be successful in close cooperation between computer science and the respective disciplines.
Impact
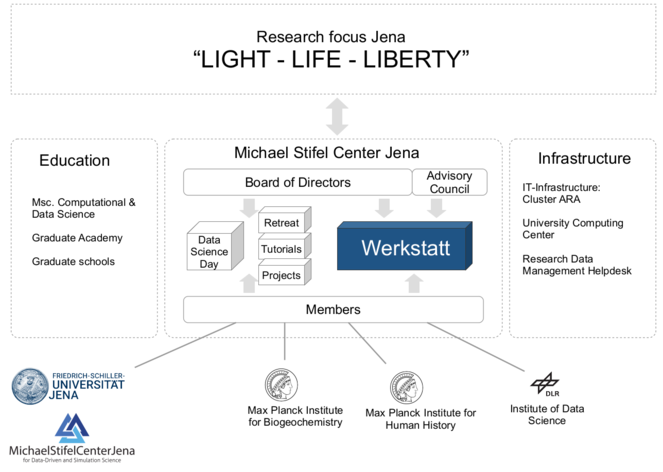
Digitization and data-driven sciences have very high priority in the strategic planning of the FSU Jena. Research on intelligent systems is of importance for all three research foci, called "profile lines'" of the FSU "Light, Life, Liberty". As early as 2015, the MSCJ was established as a cross-sectional structure. The MSCJ consists of scientists from seven faculties and three non-university research institutes in Jena (Max Planck Institute for Biogeochemistry, Max Planck Institute for Human History, DLR Institute for Data Science). This project aims to contribute to the strengthening of this structure through the development of the Werkstatt, which will facilitate cooperation both between different areas within computer science and specialist sciences, across organizations and across all career stages.
The structural goal of the project is the networking and expansion of the relevant expertise, the establishment of know-how on intelligent systems in as many specialist disciplines as possible, and the establishment of stable cooperations, thereby opening up the possibility of initiating further third-party funded projects.
Implementation
The Werkstatt comprises three core projects and ten pilot projects based on the objective areas. It also comprises activities for international networking and local communications as well as promotion of young talents. In each of the three subject areas, a five years core project is being conducted, each with one postdoctoral researcher. The three core projects shall ensure methodological continuity and also support the pilot projects with expertise and tools. The pilot projects run for three years; each is equipped with one PhD student. There will be a second, somewhat smaller set of pilot projects starting in year three of the Werkstatt. Table
Project overview and coverage of the four main themes: T1. Generic tools for machine learning, T2. Knowledge generation using machine learning processes, T3. Semantic methods for the data life cycle, and T4. Applications (black/empty square indicates primary/secondary contribution).
| Project | T1 | T2 | T3 | T4 |
|
Core Project C1: Automatic machine learning |
⬛ | ▢ | ||
|
Pilot Project P1: Probabilistic modeling |
⬛ | ▢ | ||
|
Core Project C2: Generative models |
⬛ | ▢ | ▢ | |
|
Pilot Project P2: Detection of causal relationships using deep learning |
⬛ | ▢ | ||
|
Pilot Project P3: Virus diagnostics I (Methods) |
⬛ | ▢ | ||
|
Core Project C3: Provenance management |
⬛ | ▢ | ||
|
Pilot Project P4: Data curation |
⬛ | ▢ | ||
|
Pilot Project P5: Annotation |
▢ | ⬛ | ▢ | |
|
Pilot Project P6: Gravitational waves |
▢ | ▢ | ⬛ | |
|
Pilot Project P7: Head and neck cancer |
▢ | ⬛ | ||
|
Pilot Project P8: MRI data |
⬛ | |||
|
Pilot Project P9: Phenology |
▢ | ▢ | ⬛ | |
|
Pilot Project P10: Virus diagnostics II (applications) |
▢ | ⬛ |
C1: Automatic machine learning
In order to use today’s powerful machine learning systems even without detailed machine learning knowledge, e.g., the meaning and effects of the involved hyperparameters, tools for automating the entire process of data analysis and machine learning are necessary. At the very least, one would like to assist a human analyst with the help of machine tools to process the individual steps. The aim of this core project is the further development of methods from the area of AutoML. AutoML is looking for automatic or semi-automatic methods for (1.) data preparation and transformation, (2.) selection of suitable analysis algorithms, (3.) execution of the algorithm, (4.) determination of good hyperparameters of the machine learning algorithms, (5.) selection of measures of success, (6.) data collection of missing data, and (7.) report generation.
C2: Generative models
Deep learning methods achieve their remarkable performance from a very large amount of annotated training data. However, in many applications in the sciences, e.g. in medicine, the annotation by the user/expert is very expensive or completely impossible. This core project therefore deals with modern, unsupervised learning methods and their application in the sciences. Specifically, methods for learning generative models, such as e.g. Generative Adversarial Networks (
C3: Integrated provenance management
One of the essential requirements of scientific experiments is that they are reproducible. For the reproducibility of results, it is important that the experiments are described in a structured way which contains information about the experimental setup and procedure. In recent years, a large number of domain ontologies have been created in numerous application disciplines, which formally model sections of the respective field (see, for example, the NCBI BioPortal with almost 700 ontologies). The expectation is that with the help of these ontologies, finding and linking data can be significantly improved (
The tools developed in these three core projects will be applied in the following ten pilot projects:
P1: Probabilistic modeling
In this project, new generic algorithms for learning multivariate probabilistic models are to be developed. To this end, the convex programming approach of
P2: Detection of causal relationships using deep learning
Understanding causal effects in dynamic systems plays an important role in numerous disciplines such as economy (
P3: Data-driven virus diagnostics at multiple levels I (Methods)
This project aims to develop new data analysis and learning methods to improve the diagnosis of known and unknown viruses and their interaction with other pathogens. We start from three complex data streams: sequences (genome and transcriptome of virus and host), images (virus, through TERS technology newly developed in Jena), and molecular spectra (mass spectra of virus and host), which are always cheaper and available in large quantities. We will develop new methods of machine learning and stochastic inference for the metabolomics of viral infections (molecular level). The development is based on our methods (SIRIUS, CSI: FingerID) for gas chromatography and electron ionization. We will develop learning and classification methods that use knowledge of the biological system (virus/ host) in the form of dynamic reaction networks (network level). The learning should take place both by adapting kinetic parameters and by adapting (learning) the network structure through a new form of genetic programming (CMAES-GEGP). At the molecular level and network level, knowledge is generated in the form of learned models, which should then be used for diagnostics.
P4: Data value oriented curation processes
The creation and continuous update of large knowledge bases (mainly as knowledge graphs or factual databases) is a common scenario in various disciplines, like Digital Humanities (
P5: Learning of data annotations
In many scientific disciplines, sharing data and reusing it in different contexts becomes more and more important. In order for such re-use to be truly successful, a good, ideally machine-readable description of the data is essential. However, today, creating such descriptions requires considerable manual effort. This project aims to utilize and extend machine learning techniques in creating the semantic annotations for existing datasets thus considerably reducing effort and enabling the reuse of untapped resources. Different data sources from the Biodiversity domain will be processed, with the ultimate goal to expose them as a semantic knowledge graph. To build this graph, we aim to combine different sources of information about any given dataset including the (tabular) dataset itself, existing metadata and abstracts or full texts of associated publications.
P6: Deep learning for data analysis in gravitational wave astronomy
The so-called Phenom-models (
The goal is to combine the current wave templates for concrete physical problems with new machine learning methods for data analysis. Since this field of research has only just started to attract international attention (
P7: Combined analysis of image data of head and neck cancer
Large multimodal image datasets accumulate before, during, and after therapy of head and neck cancer. The data mainly arise from endoscopic and microscopic images gained during surgery, histopathological images from tissue samples, proteomic data from MALDI imaging, as well as pre-therapeutical and post-therapeutical radiological imaging data used for tumor staging and therapy control (computed tomography, positron emission tomography, magnetic resonance imaging). So far, these different image data sources are not linked to each other, although it has been shown that the combined analysis of such data allows a much better prediction of tumor control and outcome (
P8: Use and reuse of MRI data in biomedical research environment
Assessing the data quality of biomedical imaging data, and particularly MR imaging data, is an essential prerequisite for the preparation and application of big data methods in clinical science. Conventional approaches for evaluating the data quality of magnetic resonance imaging (MRI) data are largely limited to certain sub-aspects and are often contrast-specific (
P9: Development, digitization and establishment of sensor-based phenological observations
Monitoring biodiversity is one of the major tasks in the light of global change and was identified as such in the coalition agreement of the German government. Changes in phenology which are also referred to as “fingerprint of climate change” (
The development of digitized, species-specific monitoring of phenological data in different ecosystems using sensors linked to automated image-analysis can be seen as a next step to objectively collect extensive data on plant phenology (BigData). This is not only important in the light of global change, but also offers the possibility of species-specific, automated monitoring in areas such as remote sensing and applied ecology.
P10: Data-driven virus diagnostics at multiple levels II (application)
In this project, modern analysis and learning methods are applied to achieve a breakthrough in virus diagnostics. It forms a unit with P3 (methods). On the experimental side, a fluidic chip system (artificial blood vessel) is used to infect host cells with influenza A viruses and secondarily with a pathogenic bacterium (MRSA) in a controlled manner. Furthermore, patient samples (urine, sputum, blood, BAL, CSF) from the University Hospital Jena are available and we have access to the CAPNETZ biobank with about 8000 serum samples. In the AG Marz new methods of real-time sequencing will be developed and applied (sequence level).
Real-time detection will be achieved by the MinION system (Oxford Nanopore Technologies) recently established in Jena. The system has the dimensions of a USB stick and can also be used directly on the patient. The sequencing of host DNA/RNA will be automatically stopped in real-time and another DNA/RNA fragment will be sequenced instead. The sequencing platform has already been established and tested. The tip-enhanced Raman scattering (TERS) will be used for pathogen detection in complex samples (image plane). The group in Jena has already successfully detected viruses using TERS under defined conditions. Here, protocols are being developed that allow the detection of pathogens in complex patient samples. A combination of dark-field and AFM methods ensures a fast pre-characterization of the sample, which simplifies the actual TERS characterization. As a result, we expect that the new sequence level (P10), image level (P10), molecular level (P3) and network level (P3) methods will contribute to a significant improvement in individualized real-time diagnostics.
Acknowledgements
The authors thank the Carl Zeiss Foundation for the financial support within the scope of the program line "Breakthroughs: Exploring Intelligent Systems'' for "Digitization - explore the basics, use applications''.
Funding program
Breakthroughs
Grant title
A Virtual Werkstatt for Digitization in the Sciences
Hosting institution
Friedrich Schiller University Jena, Germany
References
-
Properties of the binary black hole merger GW150914.Physical Review Letters116:241102. https://doi.org/10.1103/PhysRevLett.116.241102
-
Inspiral-merger-ringdown waveforms for black-hole binaries with non-precessing spins.Physical Review Letters106:241101. https://doi.org/10.1103/PhysRevLett.106.241101
-
Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma.Acta Oncologica56(11):1531‑1536. https://doi.org/10.1080/0284186x.2017.1346382
-
Post-radiochemotherapy PET radiomics in head and neck cancer – The influence of radiomics implementation on the reproducibility of local control tumor models.Radiotherapy and Oncology125(3):385‑391. https://doi.org/10.1016/j.radonc.2017.10.023
-
Next steps for citizen science.Science343(6178):1436‑1437. https://doi.org/10.1126/science.1251554
-
Latent variable graphical model selection via convex optimization.The Annals of Statistics40(4):1935‑1967. https://doi.org/10.1214/11-AOS949
-
Rejoinder: Latent variable graphical model selection via convex optimization.The Annals of Statistics40(4):2005‑2013. https://doi.org/10.1214/12-AOS1020
-
Using Crunchbase for economic and managerial research.OECD Science, Technology and Industry Working Papershttps://doi.org/10.1787/6c418d60-en
-
Deep neural networks to enable real-time multimessenger astrophysics.Physical Review D97(4).
-
Generative adversarial nets.Proceedings of the 27th International Conference on Neural Information Processing Systems. Volume 2.
-
Investigating causal relations by econometric models and cross-spectral methods.Econometrica37(3):424‑438. https://doi.org/10.2307/1912791
-
Improvement of olfactory function after sinus surgery correlates with white matter properties measured by diffusion tensor imaging.Neuroscience360:190‑196. https://doi.org/10.1016/j.neuroscience.2017.07.070
-
Knowledge graphs in the libraries and digital humanities domain.Encyclopedia of Big Data Technologies1‑8. https://doi.org/10.1007/978-3-319-63962-8_291-1
-
Integrated semantic search on structured and unstructured data in the ADOnIS system.Proceedings of the 2nd International Workshop on Semantics for Biodiversity co-located with 16th International Semantic Web Conference (ISWC 2017).
-
PROV-O: The PROV Ontology, 2013.W3C RecommendationURL: http://www.w3.org/TR/2013/REC-prov-o-20130430
-
Emerging problems of data quality in citizen science.Conservation Biology30(3):447‑449. https://doi.org/10.1111/cobi.12706
-
Topic maps in the eHumanities.2009 Fifth IEEE International Conference on e-Sciencehttps://doi.org/10.1109/e-science.2009.9
-
Growing season extended in Europe.Nature397(6721):659‑659. https://doi.org/10.1038/17709
-
Global warming and flowering times in Thoreau's concord: a community perspective.Ecology89(2):332‑341. https://doi.org/10.1890/07-0068.1
-
Harmonizing, annotating and sharing data in biodiversity-ecosystem functioning research.Methods in Ecology and Evolution4(2):201‑205. https://doi.org/10.1111/2041-210x.12009
-
MRI-compatible pipeline for three-dimensional MALDI imaging mass spectrometry using PAXgene fixation.Journal of Proteomics90:52‑60. https://doi.org/10.1016/j.jprot.2013.03.013
-
A globally coherent fingerprint of climate change impacts across natural systems.Nature421(6918):37‑42. https://doi.org/10.1038/nature01286
-
Causality: Models, reasoning and inference.Edition 2nd.Cambridge University Press,New York, NY, USA. https://doi.org/10.1017/CBO9780511803161
-
Elements of causal inference - foundations and learning algorithms.The MIT Press,Cambridge, MA, USA.
-
Continuous data quality assessment in information systems.Leipzig University
-
Quantitative susceptibility mapping: Concepts and applications.Clinical Neuroradiology25:225‑230. https://doi.org/10.1007/s00062-015-0432-9
-
Fingerprints of global warming on wild animals and plants.Nature421(6918):57‑60. https://doi.org/10.1038/nature01333
-
Inferring causation from time series in Earth system sciences.Nature Communications10(1):2553. https://doi.org/10.1038/s41467-019-10105-3
-
The Story of an experiment: a provenance-based semantic approach towards research reproducibility.Proceedings of the 11th International Conference Semantic Web Applications and Tools for Life Sciences, SWAT4LS 2018, Antwerp, Belgium, December 3-6, 2018.
-
Declarative metadata management: A missing piece in end-to-end machine learning.Proceedings of SYSML’18, Feb 2018, Stanford, USA.
-
The impact of over 80 years of land cover changes on bee and wasp pollinator communities in England.Proceedings of the Royal Society B: Biological Sciences282(1806):20150294‑20150294. https://doi.org/10.1098/rspb.2015.0294
-
Granger causality analysis in neuroscience and neuroimaging.Journal of Neuroscience35(8):3293‑3297. https://doi.org/10.1523/JNEUROSCI.4399-14.2015
-
Time-frequency causal inference uncovers anomalous events in environmental systems.Lecture Notes in Computer Science499‑512. https://doi.org/10.1007/978-3-030-33676-9_35
-
Denoising gravitational waves using deep learning with recurrent denoising autoencoders.arXiv: 1711.09919https://doi.org/10.1109/ICASSP.2019.8683061
-
DeepFace: closing the gap to human-level performance in face verification.Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition. https://doi.org/10.1109/CVPR.2014.220
-
Nonlinear causal link estimation under hidden confounding with an application to time series anomaly detection.Lecture Notes in Computer Science261‑273. https://doi.org/10.1007/978-3-030-33676-9_18
-
LAB-QA2GO: A free, easy-to-use toolbox for the quality assessment of magnetic resonance imaging data.Frontiers in Neuroscience13:688. https://doi.org/10.3389/fnins.2019.00688
-
MALDI-Imaging: What can be expected?European Journal of Radiology81:183‑184. https://doi.org/10.1016/s0720-048x(12)70075-x
-
Semantics in support of biodiversity knowledge discovery: an introduction to the biological collections ontology and related ontologies.PLOS One9(3):e89606. https://doi.org/10.1371/journal.pone.0089606
“Werkstatt” is the German term for “workshop” (in the sense of manufactory)