|
Research Ideas and Outcomes :
Grant Proposal
|
|
Corresponding author: Anna Heintz-Buschart (anna.heintz-buschart@ufz.de)
Received: 19 Mar 2020 | Published: 27 Mar 2020
© 2020 Anna Heintz-Buschart, Carlos Guerra, Ika Djukic, Simone Cesarz, Antonis Chatzinotas, Guillaume Patoine, Johannes Sikorski, Francois Buscot, Kirsten Küsel, Carl-Eric Wegner, Nico Eisenhauer
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Heintz-Buschart A, Guerra C, Djukic I, Cesarz S, Chatzinotas A, Patoine G, Sikorski J, Buscot F, Küsel K, Wegner C-E, Eisenhauer N (2020) Microbial diversity-ecosystem function relationships across environmental gradients. Research Ideas and Outcomes 6: e52217. https://doi.org/10.3897/rio.6.e52217
|
|
Abstract
In light of increasing anthropogenic pressures on ecosystems around the globe, the question how biodiversity change of organisms in the critical zone between Earth’s canopies and bedrock relates to ecosystem functions is an urgent issue, as human life relies on these functions. Particularly, soils play vital roles in nutrient cycling, promotion of plant growth, water purification, litter decomposition, and carbon storage, thereby securing food and water resources and stabilizing the climate. Soil functions are carried to a large part by complex communities of microorganisms, such as bacteria, archaea, fungi and protists. The assessment of microbial diversity and the microbiome's functional potential continues to pose significant challenges. Next generation sequencing offers some of the most promising tools to help shedding light on microbial diversity-function relationships. Studies relating microbial diversity and ecosystem functions are rare, particularly those on how this relationship is influenced by environmental gradients. The proposed project focuses on decomposition as one of the most important microbial soil ecosystem functions. The researchers from the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig combine an unparalleled range of expertise from next generation sequencing- based analysis of microbial communities (“meta-omics”) to soil ecology and biodiversity-ecosystem function research. This consortium will make use of soil samples from large international networks to assess microbial diversity both at the taxonomic and functional level and across the domains of life. By linking microbial diversity to functional measurements of decomposition and environmental gradients, the proposed project aims to achieve a comprehensive scale-independent understanding of environmental drivers and anthropogenic effects on the structural and functional diversity of microbial communities and subsequent consequences for ecosystem functioning.
Keywords
Biodiversity research, ecosystem functions, microbial ecology, decomposition, nutrient cycling
List of participants
Funded applicants: François Buscot, Nico Eisenhauer, Anna Heintz-Buschart, Kirsten Küsel, Carl-Eric Wegner
Co-applicants with project responsibility: Simone Cesarz, Antonic Chatzinotas, Carlos Guerra, Johannes Sikorski
International cooperation partner: Ika Djukic
Further project group: Guillaume Pantoine, Tesfaye Wubet
Third parties involved in the project
The project is performed in cooperation with international partners. Ika Djukic (Swiss Federal Institute for Forest, Snow and Landscape Research WSL, Zürcherstrasse 111, 8903 Birmensdorf; - Switzerland; ika.djukic@umweltbundesamt.at) coordinates the global efforts of TeaComposition, the soil samples of which will be analysed in this project. Ika Djukic is not financially involved beyond the provision of the teabags to the cooperating scientists. There is no financial involvement in the proposed analyses for either partner.
Next generation sequencing will be performed at a DFG Sequencing Centre, the NGS Competence Center Tübingen (NCCT), Germany.
State of the art and preliminary work
Human activities are altering the biodiversity of ecosystems around the globe, which has provoked concern regarding potential consequences for ecosystem functions. The notion that biodiversity could be an important determinant of ecosystem functions stems from observations of natural communities (
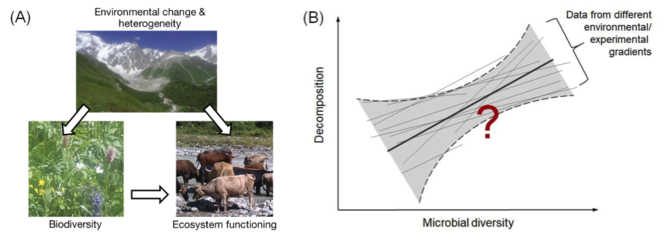
A Environmental change and heterogeneity determine the biodiversity and the functioning of ecosystems (modified after
Future approaches to study BEF relationships should thus move from studying the relationships between random species loss and ecosystem functioning to the complex interplay between anthropogenic drivers, non-random biodiversity changes (including gains, losses, and changes in evenness/dominance and composition/traits), ecosystem functioning, and ecosystem services at multiple spatial and temporal scales. Given these complications, evidence for significant BEF relationships across environmental gradients is scarce (e.g.,
Currently, most BEF studies focus on aboveground diversity, although the ecosystem functions are driven by belowground biological processes. In particular, soils play pivotal roles in important terrestrial ecosystem functions including nutrient cycling, sustaining plant growth, water purification, litter decomposition, and carbon storage (
Although microcosm experiments suggest positive microbial BEF relationships (e.g.,
Among the important functions fulfilled by soil microbial communities that are strongly influenced by soil conditions, climate, and human activities (e.g.,
We lack a systemic understanding of how this cycling potential is influenced by local (e.g. along soil profile) and interregional variations (climate, soil type, stoichiometry) via their impacts on soil biodiversity. For the longest time, global litter decomposition has been considered to be mainly controlled by climate (temperature and moisture) and litter quality (i.e. the chemical composition;
With the proposed project, we aim to perform an integrative, global assessment of the relationship between microbial diversity and litter decomposition in the soil, using a set of existing monitoring platforms operating along environmental gradients (Fig.
Given the strong societal relevance of global soil biodiversity and function (
Moreover, some of the PIs have been active participants in the TeaComposition network (
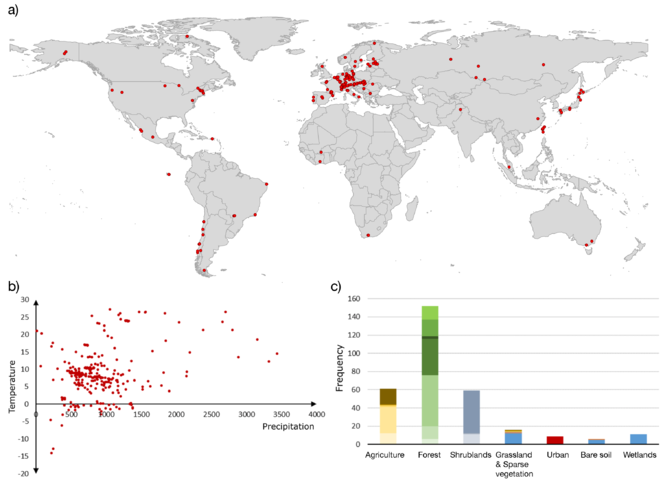
Overview of the sample collection. Decomposition, as a central soil function, and the biodiversity of involved microbial communities will be studied in a large, international monitoring network (sampling locations indicated in a) to assess the generality of microbial biodiversity-function relationships across climates (mean annual air temperature and annual precipitation in b) and land use and cover types (c).
In summary, this project will foster integration between the above-mentioned approaches to reach a comprehensive scale-independent understanding of environmental drivers and anthropogenic effects on the structural and functional diversity of microbial communities and the subsequent consequences for ecosystem functioning. Therefore, we will produce extensive datasets on the main soil microbial taxa (bacteria, archaea, fungi, and protists) across a global network of sites in relation to climate and land use. Functional relationships will be deduced by multivariate analyses (e.g., structural equation modelling;
Project-related publications
- Reich PB, Tilman D, Isbell F, Mueller K, Hobbie SE, Flynn DFB and Eisenhauer N (2012) Impacts of biodiversity loss escalate through time as redundancy fades. Science 336: 589-592.
- Isbell F, Craven D, Connolly J, Loreau M, Schmid B, Beierkuhnlein H, Bezemer TM, Bonin C, Bruelheide H, de Luca E, Ebeling A, Griffin J, Guo Q, Hautier Y, Hector A, Jentsch A, Kreyling J, Lanta V, Manning P, Meyer ST, Mori AS, Naeem S, Niklaus PA, Polley HW, Reich PB, Roscher C, Seabloom EW, Smith MD, Thakur MP, Tilman D, Tracy BF, van der Putten WH, van Ruijven J, Weigelt A, Weisser WW, Wilsey B, Eisenhauer N (2015) Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 526: 574-577.
- Roume H, Heintz-Buschart A, Muller EEL, May P, Satagopam VP, Laczny CC, Narayanasamy S, Lebrun L, Hoopmann MR, Schupp JM, Gillece JD, Hicks ND, Engelthaler DM, Sauter T, Keim PS, Moritz RL, Wilmes P (2015) Comparative integrated omics: identification of key functionalities in microbial community-wide metabolic networks. NPJ Biofilms and Microbiomes 1: 15007.
- European Commission, Joint Research Centre. 2016. Global Soil Biodiversity Atlas - Earth Sciences Research - EU Bookshop. bookshop.europa.eu/en/global-soil-biodiversity-atlas-pbLBNA27236/. Contributors: Küsel K, Totsche KU, Trumbore SE, Buscot F; Chapter III - Geographical and temporal distribution, p. 73.
- Ivanova AA, Wegner CE, Kim Y, Liesack W, Dedysh SN. (2016). Identification of microbial populations driving biopolymer degradation in acidic peatlands by metatranscriptomic analysis. Molecular Ecology 25: 4818-4835.
- Beulig F, Urich T, Nowak M, Trumbore SE, Gleixner G, Gilfillan GD, Fjelland KE, Küsel K 2016. Altered carbon turnover processes and microbiomes in soils under long-term extremely high CO2 exposure. Nature Microbiology 1: 15025.
- Purahong W, Wubet T, Lentendu G, Schloter M, Pecyna MJ, Kapturska D, Hofrichter M, Kruger D, Buscot F (2016) Life in leaf litter: novel insights into community dynamics of bacteria and fungi during litter decomposition. Molecular Ecology 25: 4059-4074.
- Soliveres S, van der Plas F, Manning P, Prati D, Gossner MM, Renner SC, Alt F, Arndt H, Baumgartner V, Binkenstein J, Birkhofer K, Blaser S, Blüthgen N, Boch S, Böhm S, Börschig C, Buscot F, Diekötter T, Heinze J, Hölzel N, Jung K, Klaus VH, Kleinbecker T, Klemmer S, Krauss J, Lange M, Morris EK, Müller J, Oelmann Y, Overmann J, Pašalić E, Rillig MC, Schaefer M, Schloter M, Schmitt B, Schöning I, Schrumpf M, Sikorski J, Socher SA, Solly EF, Sonnemann L, Sorkau E, Steckel J, Steffan-Dewenter I, Stempfhuber B, Tschapka M, Türke M, Venter PC, Weiner CN, Weisser WW, Werner M, Westphal C, Wilcke W, Wolters V, Wubet T, Wurst S, Fischer M, Allan E (2016) Biodiversity at multiple trophic levels is needed for ecosystem multifunctionality. Nature 536: 456-459.
- Wegner CE, Liesack W (2016) Microbial community dynamics during the early stages of plant polymer breakdown in paddy soil. Environmental Microbiology 18; 2825-2842.
- Guerrero-Ramirez NR, Craven D, Reich PB, Ewel JJ, Isbell F, Koricheva J, Parrotta JA, Auge H, Erickson HE, Forrester DI, Hector H, Joshi J, Montagnini F, Palmborg C, Piotto D, Potvin C, Roscher C, van Ruijven J, Tilman D, Wilsey B and Eisenhauer N (2017) Diversity-dependent temporal divergence of ecosystem functioning in experimental ecosystems. Nature Ecology & Evolution 1: 1639-1642.
Objectives and work programm
Anticipated total duration of the project
iDiv is a Research Centre of the DFG, established in 2012, currently in the second funding phase (October 2016 - September 2020). The integrative project “Microbial diversity-ecosystem function relationships across environmental gradients” described here runs since November 2017, with monthly meetings, and supported by an iDiv postdoc (Dr. Carlos Guerra) since June 2018. The DFG-funded NGS applied for in this proposal should take place in the first half of 2020 (see Table
Project duration: NGS should be performed in the first half of 2020.
| 2017-2019 |
2020 II |
2020 III |
2020 IV |
2021 I |
2021 II |
|
| Sample collection | x | |||||
| Interaction with NGS centre | x | ship samples &receive data | ||||
| Data processing amplicons | x | |||||
| Data processing metagenomics | x | x | ||||
| Integration | x | x | ||||
| Publication | x |
Objectives
This project aims to explore the relationship between microbial diversity and decomposition, as a major ecosystem function carried out by microbes of the soil, across environmental contexts (Fig.
Despite the global origin of the samples contributed by the TeaComposition network, there are several unifying characteristics, which will enable integration of the results. To investigate the direct effects of different environmental drivers on soil biodiversity and related functions, the network analyses a standardized decomposing material. Sampling procedures have been synchronized, and the same sequencing approaches will be used for all samples. Decomposition rates at all sites after 3 months, 1 year, and 2 years have been determined in the framework of TeaComposition (
This study makes use of a network of decomposition experiments across the globe, with more than 350 participating sites (Fig.
Although human activities have accelerated global biodiversity change and have threatened the supply of ecosystem services, global biodiversity assessments, syntheses, and predictions are based primarily on a few well-studied aboveground taxa including plants and vertebrates (e.g., Millennium Ecosystem Assessment; IUCN red lists;
- the drivers of soil microbial diversity, and
- the consequences for important functions.
In this context, the NGS data will directly contribute to inform about the global and local relations between soil biodiversity and decomposition. By doing this at multiple scales, it will also support the development of complex causal models that will integrate the global model intercomparison project for soils [Soil-MIP, https://soil-modeling.org/science-panels/Soil-MIP] and will be linked to global soil biodiversity assessments. The NGS data will be used in at least one integrative publication in a high-ranking journal and a data descriptor. In addition, the data will greatly advance the global soil biodiversity database [EDAPHOBASE, https://portal.edaphobase.org], and support the global biodiversity network of the Group in Earth Observations (www.geobon.org).
Work programme incl. proposed research methods
The presented project is a collaborative effort with contributions of each of the PIs. In an integrative effort, all key soil microbial taxa (bacteria, fungi, archaea, and protists) will be comprehensively profiled in a worldwide collection of functionally characterised soil samples by the collaborating groups using amplicon sequencing. To establish a link between the soil microbial molecular functional capacity and ecosystem functions, the samples will be further analysed using shotgun metagenomics, yielding information on functional diversity and distribution patterns of molecular functions related to decomposition. The resulting unprecedented dataset will allow us to study relationships between microbial diversity and ecosystem functions as well as ecosystem multifunctionality.
We will synthesize data on microbial diversity-ecosystem function relationships across the target taxa and functions to develop a scale-independent understanding of environmental drivers and anthropogenic effects on the structural and functional diversity of microbial communities and the subsequent consequences for ecosystem functioning (Fig.
In particular, the tasks shared by the project partners are:
- communication with NGS centre and DFG, coordination of sample shipping (DNA from 2 x 395 soil samples collected worldwide within the TeaComposition network (
Djukic et al. 2018 ), presently stored at iDiv) (A. Heintz-Buschart) - devising and implementing the data management plan, data transfer from NGS centres and sharing among consortium (A. Heintz-Buschart)
- coordination of NGS data processing, bioinformatics support (A. Heintz-Buschart)
- analysis of prokaryotic community structure and diversity of 800 soil samples (K. Küsel, in collaboration with T. Wubet and J. Sikorski)
- analysis of eukaryotic community structure and diversity of 800 soil samples (F. Buscot, in collaboration with A. Chatzinotas)
- analysis of functional diversity in 395 soil samples (A. Heintz-Buschart, in collaboration with K. Küsel and C.-E. Wegner)
- analysis of occurrence of decomposition-related genes in 395 soil samples (C.-E. Wegner, in collaboration with K. Küsel and A. Heintz-Buschart)
- relating decomposition data to data on soil microbial biomass, microbial respiration, soil aggregate stability (all generated in the Eisenhauer lab), and microbial community composition (N. Eisenhauer in collaboration with S. Cesarz and I. Djukic)
- to study microbial diversity-function relationships across large environmental gradients (N. Eisenhauer in collaboration with C. Guerra and S. Cesarz)
- to perform microbial diversity-function analyses related to litter decomposition across large environmental gradients (N. Eisenhauer in collaboration with C. Guerra, I. Djukic, J. Sikorski and A. Heintz-Buschart)
- preparation and documentation of meetings of project consortium (A. Heintz-Buschart in collaboration with C. Guerra)
- dissemination of results (all project partners)
Project background
This project will be carried out within the TeaComposition network, taking advantage of a globally distributed research infrastructure on decomposition of organic matter and aims at providing a more comprehensive understanding of biosphere-atmosphere carbon feedback. It is a global low cost and “easy-to-join” initiative that is open for international collaboration. In addition to the global sampling approach, the research conducted in this network will provide an improved understanding of decomposition for leading experiments and observation networks, such as ILTER, CLIMMANI, TreeDivNet, and GLORIA. Decomposition trials have already been analysed (12 and 24 months litter mass loss) and published (three months litter mass loss;
Sampling design and soil data
This study uses experiments and observation networks at more than 350 sites worldwide (Fig. 3a). Each site consists of 2 blocks, with 2 tea bag types that represent different litter qualities (
Sequencing requirements
Both blocks of all 395 sites will be analysed by amplicon sequencing (790 samples in total). Deep microbial community profiles will be generated based on four regions within the rRNA operons, that are specifically informative for the major microbial taxa in soil, namely 16S V4 for bacteria and many archaea, ITS for fungi and two 18S regions for fungi and protists. A total of 400,000 read pairs per sample are required. In addition, shotgun metagenomics of one of the blocks in each site will be performed to derive measures of functional diversity and to survey the occurrence of genes involved in decomposition, such as carbohydrate-active enzymes. We estimate a depth of 50 million read pairs per sample to be informative for this aim.
Sequencing data analysis
Amplicon sequencing data will be processed separately for each target using established pipelines based on frameworks such as Mothur (
Whole shotgun metagenomic data will be processed using a pipeline already set up on the high performance computing infrastructure at the iDiv, starting from quality filtering and de novo assembly (
Together, the amplicon and shotgun data will yield deeply sampled measures of bacterial, archaean, fungal and protist taxonomic and phylogenetic diversity, complemented by measures of global and decomposition-specific genetic functional diversity of the more abundant taxa.
Together, the amplicon and shotgun data will yield deeply sampled measures of bacterial, archaean, fungal and protist taxonomic and phylogenetic diversity, complemented by measures of global and decomposition-specific genetic functional diversity of the more abundant taxa.
Data integration
We will test if significant BEF relationships across environmental gradients exist, in particular with respect to microbial diversity measures and decomposition. We expect an overall positive relationship with heterogeneous slopes in diversity gradients caused by different environmental drivers (Fig.
Like done in previous synthesis projects (e.g.
Data handling
This project will integrate existing data, as detailed above, with data sets to be derived from NGS. In the long-term, the NGS data pertaining to specific geographic locations will remain of value to the research community. The publication of the datasets are not restricted by privacy concerns. Samples and data are subject to the CBD (see section 2.5).
The data management plan for this project will be devised and implemented by Anna Heintz-Buschart in consolation with all project partners, and if necessary supported by the Biodiversity Informatics Unit of iDiv. The following measures are foreseen in order to make the generated data available for future re-use: The NGS data will be made publicly available at the time of the publication of results, or at the very latest at the end of the project, in international public databases in openly documented standard data formats. In these databases, the data will be findable, publically accessible, linked to the databases hosting other types of data from the project, together with data descriptors in standard formats, and therefore the data will be interoperable and reusable (according to the FAIR - findable, accessible, interoperable, reusable - principles). The publication of a data descriptor in an open access journal such as Scientific Data or Giga Science is foreseen. Raw NGS reads will be submitted to the NCBI’s short read archive or the European Nucleotide Archive ENA, which links them to geo-referenced ‘biosample’ data handles that will contain also soil and plant residue physicochemical parameters and sample metadata. Environmental and ecosystem function data will also be submitted and cross-linked within the PANGAEA database and have a DOI of the iDiv data portal (https://idata.idiv.de). Derived data types such as reconstructed genomes will likewise be submitted to dedicated, open databases fulfilling Minimal Information standards. All computational workflows used to process the NGS data will be linked to the published datasets and will be open source, in order to ensure reproducible bioinformatics. They will be kept in sustainable repositories, such as CERN’s Zenodo, where they are accessible via a DOI. The data management plan foresees that all long-term data will have been submitted to public databases by the end of the project and no long-term costs will be incurred for the consortium.
Other information
The NGS analyses are subject to the Convention on Biological Diversity (CBD). The applicants are familiar with the CBD and the DFG’s guidelines on the CBD. The applicants will ensure that all analyses will conform to the rules of the CBD. As no use beyond basic research and no commercialization are envisaged, a simple ABS contracts will be sufficient.
Material transfer agreement with the collaborating scientist abroad (and in Germany) have been signed. The applicants have localised the national contact points and inquired for the processes in the participating countries. We are optimistic that PICs and MATs can be achieved for the remaining countries. To comply with all legal regulations, the applicants receive support by “Dezernat für Forschungs- und Transferservice” of Leipzig University.
Budget: Requested modules/funds
This project proposal is to request funding of NGS analyses, as detailed in section 2 of this proposal. Sufficient funding for all other costs are available to the applicants within the framework of iDiv. Therefore, no funding is requested for any of the following points:
Funding for Staff; Direct Project Costs; Equipment up to 10 000 €, Software and Consumables; Travel Expenses; Visiting Researchers; Expenses for Laboratory Animals; Project-related publication expenses; Instrumentation; Equipment exceeding 10 000 €; Major Instrumentation exceeding 50 000 €.
Other costs: NGS costs
This proposal is for NGS costs of an on-going project within the DFG Research Centre iDiv, which is not covered by the current iDiv business plan or any of the experimental platforms and networks. All costs beside the NGS costs will be carried by the applicants and iDiv funding. The proposed project will incur sequencing costs of 202 172 €, as detailed in the quotations and Counselling Reports from the DFG NGS centre in Tübingen. Table
Budget.
|
Method |
Samples |
Targets |
Costs |
cost details |
|
Amplicon sequencing |
800 |
4 |
75 241 € |
12.78 € × 3200 = 40 911 €
34 330 € |
|
Metagenomics |
395 |
1 |
121 561 € |
66.08 € × 395 = 26 101 €
15 805 € × 6 = 100 830 € |
|
total |
202 172 € |
|||
|
per PI |
40 434 € |
Justification: Amplicon sequencing is requested as the only reliable method to deeply profile all target soil taxa, including rare taxa, and assess inter-kingdom interactions. The proposal foresees the use of the NGS centre’s automation and high-throughput sequencers, potentially including a highly multiplexed approach to enable the high number of samples. To perform functional profiling of the prominent taxa, metagenomics measurements are requested. Both targeted and shotgun technologies are combined to compare and facilitate changes from amplicon-based to whole genome techniques if sequencing volumes and depth per € increase further in the future.
Project requirements
Applicants' employment status information
Applicants are detailed in Table
Applicants.
|
Last name |
First name |
Title |
Employment and funding body |
|
Heintz-Buschart |
Anna |
Dr. |
Bioinformatics Unit of German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, employed by Helmholtz Centre for Environmental Research GmbH – UFZ, fixed term contract, funded by the German Research Foundation - DFG (FZT 118 – iDiv) |
|
Küsel |
Kirsten |
Prof. Dr. |
Friedrich Schiller University - Institute of Biodiversity, permanent |
|
Wegner |
Carl-Eric |
Dr. |
Friedrich Schiller University - Institute of Biodiversity, fixed term contract, funded by Friedrich Schiller University |
|
Eisenhauer |
Nico |
Prof. Dr. |
Professor at German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig and Leipzig University, permanent |
|
Buscot |
François |
Prof. Dr. |
Helmholtz Centre for Environmental Research GmbH – UFZ, Soil Ecology Department, permanent |
Composition of the project group
This project will involve several researchers of iDiv and the PIs’ working groups at Friedrich-Schiller-University Jena and the UFZ contributing their experience to the experimental work packages and/or the integration, see Table
Project group.
|
Last name |
First name |
Title |
Employment, duration and funding body |
|
Guerra |
Carlos |
Dr. |
Experimental Interaction Ecology group at iDiv and Leipzig University, fixed term contract, funded by the German Research Foundation - DFG (FZT 118 – iDiv) |
|
Cesarz |
Simone |
Dr. |
Experimental Interaction Ecology group at iDiv and Leipzig University, permanent |
|
Sikorski |
Johannes |
Dr. |
Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Culture, permanent |
|
Chatzinotas |
Antonis |
Dr. |
Helmholtz Centre for Environmental Research GmbH – UFZ, permanent |
|
Wubet |
Tesfaye |
Dr. |
Helmholtz Centre for Environmental Research GmbH – UFZ, permanent |
Scientific equipment
The proposed projects make use of existing experimental platforms and computing infrastructure. For the analysis of the NGS data in all WPs, the high-performance computing cluster of UFZ and iDiv will be used.
Funding program
Research Grants
Grant title
Microbial diversity-ecosystem function relationships across environmental gradients
Hosting institution
Helmholtz-Centre for Environmental Research GmbH - UFZ, Halle, Germany
University Leipzig, Germany
Friedrich-Schiller-University Jena, Germany
Conflicts of interest
The authors have declared that no competing interests exist.
References
-
Microbial abundance and composition influence litter decomposition response to environmental change.Ecology94(3):714‑725. https://doi.org/10.1890/12-1243.1
-
Structure and function of the global topsoil microbiome.Nature560(7717):233‑237. https://doi.org/10.1038/s41586-018-0386-6
-
Belowground biodiversity and ecosystem functioning.Nature515(7528):505‑511. https://doi.org/10.1038/nature13855
-
Understanding the dominant controls on litter decomposition.Journal of Ecology104(1):229‑238. https://doi.org/10.1111/1365-2745.12507
-
DADA2: High-resolution sample inference from Illumina amplicon data.Nature Methods13(7):581‑583. https://doi.org/10.1038/nmeth.3869
-
Global gaps in soil biodiversity data.Nature Ecology & Evolution2(7):1042‑1043. https://doi.org/10.1038/s41559-018-0573-8
-
Global mismatches in aboveground and belowground biodiversity.Conservation Biology33(5):1187‑1192. https://doi.org/10.1111/cobi.13311
-
Biodiversity loss and its impact on humanity.Nature486(7401):59‑67. https://doi.org/10.1038/nature11148
-
Meta-analysis shows positive effects of plant diversity on microbial biomass and respiration.Nature Communications10:1332. https://doi.org/10.1038/s41467-019-09258-y
-
Litter decomposition, climate and liter quality.Trends in Ecology & Evolution10(2):63‑66. https://doi.org/10.1016/s0169-5347(00)88978-8
-
The unseen invaders: introduced earthworms as drivers of change in plant communities in North American forests (a meta-analysis).Global Change Biology23(3):1065‑1074. https://doi.org/10.1111/gcb.13446
-
Plant diversity effects on grassland productivity are robust to both nutrient enrichment and drought.Philosophical Transactions of the Royal Society B: Biological Sciences371(1694):20150277. https://doi.org/10.1098/rstb.2015.0277
-
Reintroducing environmental change drivers in biodiversity–ecosystem functioning research.Trends in Ecology & Evolution31(12):905‑915. https://doi.org/10.1016/j.tree.2016.09.007
-
Microbial diversity drives multifunctionality in terrestrial ecosystems.Nature Communications7:10541. https://doi.org/10.1038/ncomms10541
-
A global atlas of the dominant bacteria found in soil.Science359(6373):320‑325. https://doi.org/10.1126/science.aap9516
-
Cross-biome drivers of soil bacterial alpha diversity on a worldwide scale.Ecosystems22(6):1220‑1231. https://doi.org/10.1007/s10021-018-0333-2
-
Mediterranean grassland soil C–N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms.Nature Microbiology4(8):1356‑1367. https://doi.org/10.1038/s41564-019-0449-y
-
Early stage litter decomposition across biomes.The Science of the Total Environment628-629:1369‑1394. https://doi.org/10.1016/j.scitotenv.2018.01.012
-
Plant diversity effects on soil microorganisms support the singular hypothesis.Ecology91(2):485‑496. https://doi.org/10.1890/08-2338.1
-
Biodiversity-ecosystem function experiments reveal the mechanisms underlying the consequences of biodiversity change in real world ecosystems.Journal of Vegetation Science27(5):1061‑1070. https://doi.org/10.1111/jvs.12435
-
Plant diversity maintains multiple soil functions in future environments.eLIFE7:e41228. https://doi.org/10.7554/eLife.41228
-
Recognizing the quiet extinction of invertebrates.Nature Communications10(1). https://doi.org/10.1038/s41467-018-07916-1
-
The Pfam protein families database in 2019.Nucleic Acids Research47(D1):D427‑D432. https://doi.org/10.1093/nar/gky995
-
The ecology of invasions by animals and plants.Methuen and Co. Ltd.,London. https://doi.org/10.1007/978-1-4899-7214-9
-
Effects of plant functional group removal on structure and function of soil communities across contrasting ecosystems.Ecology Letters22:1095‑1103. https://doi.org/10.1111/ele.13266
-
Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients.The ISME Journal6(5):1007‑1017. https://doi.org/10.1038/ismej.2011.159
-
Using plant, microbe, and soil fauna traits to improve the predictive power of biogeochemical models.Methods in Ecology and Evolution10:135‑33. https://doi.org/10.1111/2041-210X.13092
-
Metatranscriptomic census of active protists in soils.The ISME Journal9(10):2178‑2190. https://doi.org/10.1038/ismej.2015.30
-
A niche for ecosystem multifunctionality in global change research.Global Change Biology25:763‑774. https://doi.org/10.1111/gcb.14528
-
Ecological assembly rules in plant communities-approaches, patterns and prospects.Biological Reviews87(1):111‑127. https://doi.org/10.1111/j.1469-185X.2011.00187.x
-
Integrative modelling reveals mechanisms linking productivity and plant species richness.Nature529(7586):390‑393. https://doi.org/10.1038/nature16524
-
Diversity-dependent temporal divergence of ecosystem functioning in experimental ecosystems.Nature Ecology & Evolution1(11):1639‑1642. https://doi.org/10.1038/s41559-017-0325-1
-
The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy.Nucleic Acids Research41(D1):D597‑D604. https://doi.org/10.1093/nar/gks1160
-
Species’ traits predict the effects of disturbance and productivity on diversity.Ecology Letters11(4):348‑356. https://doi.org/10.1111/j.1461-0248.2007.01149.x
-
Biodiversity and Litter Decomposition in Terrestrial Ecosystems.Annual Review of Ecology, Evolution, and Systematics36(1):191‑218. https://doi.org/10.1146/annurev.ecolsys.36.112904.151932
-
Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes.Nature Microbiology2:16180. https://doi.org/10.1038/nmicrobiol.2016.180
-
dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation.Nucleic Acids Research46(D1). https://doi.org/10.1093/nar/gkx894
-
eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences.Nucleic Acids Research44(D1). https://doi.org/10.1093/nar/gkv1248
-
Nutrient enrichment, biodiversity loss, and consequent declines in ecosystem productivity.Proceedings of the National Academy of Sciences of the United States of America110(29):11911‑11916. https://doi.org/10.1073/pnas.1310880110
-
Biodiversity increases the resistance of ecosystem productivity to climate extremes.Nature526(7574):574‑577. https://doi.org/10.1038/nature15374
-
The influence of soil communities on the temperature sensitivity of soil respiration.Nature Ecology & Evolution2(10):1597‑1602. https://doi.org/10.1038/s41559-018-0648-6
-
Genotypic richness and dissimilarity opposingly affect ecosystem functioning.Ecology Letters14(6):537‑545. https://doi.org/10.1111/j.1461-0248.2011.01613.x
-
Using intron position conservation for homology-based gene prediction.Nucleic Acids Research44(9). https://doi.org/10.1093/nar/gkw092
-
Disentangling the mechanisms underlying functional differences among decomposer communities.Journal of Ecology102(3):603‑609. https://doi.org/10.1111/1365-2745.12220
-
Aggregate stability and size distribution.Methods of Soil AnalysisURL: https://eprints.nwisrl.ars.usda.gov/732/3/585.pdf
-
Tea Bag Index: a novel approach to collect uniform decomposition data across ecosystems.Methods in Ecology and Evolution4(11):1070‑1075. https://doi.org/10.1111/2041-210X.12097
-
Nitrogen additions and litter decomposition: A meta‐analysis.Ecology86(12):3252‑3257. https://doi.org/10.1890/05-0150
-
Towards a unified paradigm for sequence-based identification of fungi.Molecular Ecology22(21):5271‑5277. https://doi.org/10.1111/mec.12481
-
Inference of phenotype-defining functional modules of protein families for microbial plant biomass degraders.Biotechnology for Biofuels7:124. https://doi.org/10.1186/s13068-014-0124-8
-
Plant diversity increases soil microbial activity and soil carbon storage.Nature Communications6(1). https://doi.org/10.1038/ncomms7707
-
Biodiversity enhances ecosystem multifunctionality across trophic levels and habitats.Nature Communications6(1):6707. https://doi.org/10.1038/ncomms7936
-
Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe.Proceedings of the National Academy of Sciences112(35):10967‑10972. https://doi.org/10.1073/pnas.1508382112
-
Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits.The ISME Journal12(7):1794‑1805. https://doi.org/10.1038/s41396-018-0089-x
-
Soil biota contributions to soil aggregation.Nature Ecology & Evolution1(12):1828‑1835. https://doi.org/10.1038/s41559-017-0344-y
-
The Global Stoichiometry of Litter Nitrogen Mineralization.Science321(5889):684‑686. https://doi.org/10.1126/science.1159792
-
Links between plant litter chemistry, species diversity, and below-ground ecosystem function.Proceedings of the National Academy of Sciences of the United States of America105(50):19780‑19785. https://doi.org/10.1073/pnas.0805600105
-
AnnoTree: visualization and exploration of a functionally annotated microbial tree of life.Nucleic Acids Research304:66‑7. https://doi.org/10.1093/nar/gkz246
-
The functions of biological diversity in an age of extinction.Science336(6087):1401‑1406. https://doi.org/10.1126/science.1215855
-
IMP: a pipeline for reproducible reference-independent integrated metagenomic and metatranscriptomic analyses.Genome Biology17(1).
-
The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota).New Phytologist188(1):223‑241. https://doi.org/10.1111/j.1469-8137.2010.03334.x
-
A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life.Nature Biotechnology15https://doi.org/10.1038/nbt.4229
-
Mechanisms linking plant community properties to soil aggregate stability in an experimental grassland plant diversity gradient.Plant and Soil373(1-2):285‑299. https://doi.org/10.1007/s11104-013-1791-0
-
Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide.Ecology Letters18(1):85‑95. https://doi.org/10.1111/ele.12381
-
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools.Nucleic Acids Research41(Database Issue):590‑596. https://doi.org/10.1093/nar/gks1219
-
The rational exploration of microbial diversity.The ISME Journal2(10):997‑1006. https://doi.org/10.1038/ismej.2008.69
-
Toward a global platform for linking soil biodiversity data.Frontiers in Ecology and Evolution3:91. https://doi.org/10.3389/fevo.2015.00091
-
Detecting macroecological patterns in bacterial communities across independent studies of global soils.Nature Microbiology3(2):189‑196. https://doi.org/10.1038/s41564-017-0062-x
-
Mechanisms driving the soil organic matter decomposition response to nitrogen enrichment in grassland soils.Soil Biology and Biochemistry99(C):54‑65. https://doi.org/10.1016/j.soilbio.2016.04.023
-
Abundance determines the functional role of bacterial phylotypes in complex communities.Nature Microbiology320https://doi.org/10.1038/s41564-018-0180-0
-
Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities.Applied and Environmental Microbiology75(23):7537‑7541. https://doi.org/10.1128/AEM.01541-09
-
The value of biodiversity experiments.Basic and Applied Ecology5(6):535‑542. https://doi.org/10.1016/j.baae.2004.07.001
-
Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions.The ISME Journal6(9):1749‑1762. https://doi.org/10.1038/ismej.2012.11
-
Prokka: rapid prokaryotic genome annotation.Bioinformatics30(14):2068‑2069. https://doi.org/10.1093/bioinformatics/btu153
-
Microbial Functional Diversity Associated with Plant Litter Decomposition Along a Climatic Gradient.Microbial Ecology64(2):399‑415. https://doi.org/10.1007/s00248-012-0037-7
-
Loss of microbial diversity in soils is coincident with reductions in some specialized functions.Environmental Microbiology16(8):2408‑2420. https://doi.org/10.1111/1462-2920.12353
-
Biodiversity at multiple trophic levels is needed for ecosystem multifunctionality.Nature536(7617):456‑459. https://doi.org/10.1038/nature19092
-
Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources.BMC Bioinformatics7https://doi.org/10.1186/1471-2105-7-62
-
Fungal biogeography. Global diversity and geography of soil fungi.Science346(6213):1256688. https://doi.org/10.1126/science.1256688
-
Composition of natural organic materials and their decomposition in the soil: IV. The nature and rapidity of decomposition of the various organic complexes in different plant materials, under aerobic conditions.Soil Sciences28:55. https://doi.org/10.1097/00010694-192907000-00005
-
Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training.Genome Research18(12):1979‑1990. https://doi.org/10.1101/gr.081612.108
-
Plant diversity drives soil microbial biomass carbon in grasslands irrespective of global environmental change factors.Global Change Biology21(11):4076‑4085. https://doi.org/10.1111/gcb.13011
-
Plant diversity and ecosystem productivity: theoretical considerations.Proceedings of the National Academy of Sciences94(5):1857‑1861. https://doi.org/10.1073/pnas.94.5.1857
-
Facilitative interactions rather than resource partitioning drive diversity-functioning relationships in laboratory fungal communities.Ecology Letters8(6):618‑625. https://doi.org/10.1111/j.1461-0248.2005.00757.x
-
Nitrogen additions and microbial biomass: a meta-analysis of ecosystem studies.Ecology Letters11(10):1111‑1120. https://doi.org/10.1111/j.1461-0248.2008.01230.x
-
The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems.Ecology Letters11(3):296‑310. https://doi.org/10.1111/j.1461-0248.2007.01139.x
-
A thready affair: linking fungal diversity and community dynamics to terrestrial decomposition processes.FEMS Microbiology Reviews37(4):477‑494. https://doi.org/10.1111/1574-6976.12001
-
Do arbuscular mycorrhizal fungi stabilize litter-derived carbon in soil?Journal of Ecology104(1):261‑269. https://doi.org/10.1111/1365-2745.12496
-
Soil biodiversity and human health.Nature528(7580):69‑76. https://doi.org/10.1038/nature15744
-
Do experiments exploring plant diversity-ecosystem functioning relationships inform how biodiversity loss impacts natural ecosystems?Journal of Vegetation Science27(3):646‑653. https://doi.org/10.1111/jvs.12399
-
Genome-reconstruction for eukaryotes from complex natural microbial communities.Genome Research28(4):569‑580. https://doi.org/10.1101/gr.228429.117
-
Kraken: ultrafast metagenomic sequence classification using exact alignments.Genome Biology15(3). https://doi.org/10.1186/gb-2014-15-3-r46
-
Soil protist communities form a dynamic hub in the soil microbiome.The ISME Journal12(2):634‑638. https://doi.org/10.1038/ismej.2017.171
-
Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis.Proceedings of the National Academy of Sciences96(4):1463‑1468. https://doi.org/10.1073/pnas.96.4.1463
-
Global negative effects of nitrogen deposition on soil microbes.The ISME Journal12(7):1817‑1825. https://doi.org/10.1038/s41396-018-0096-y