|
Research Ideas and Outcomes : Grant Proposal
|
|
Corresponding author: Caroline Müller (caroline.mueller@uni-bielefeld.de)
Received: 02 Jan 2020 | Published: 21 Jan 2020
© 2020 Caroline Müller, Andrea Bräutigam, Elisabeth Eilers, Robert Junker, Jörg-Peter Schnitzler, Anke Steppuhn, Sybille Unsicker, Nicole van Dam, Wolfgang Weisser, Meike Wittmann
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Müller C, Bräutigam A, Eilers EJ, Junker RR, Schnitzler J-P, Steppuhn A, Unsicker SB, van Dam NM, Weisser WW, Wittmann MJ (2020) Ecology and Evolution of Intraspecific Chemodiversity of Plants. Research Ideas and Outcomes 6: e49810. https://doi.org/10.3897/rio.6.e49810
|
|
Abstract
An extraordinarily high intraspecific chemical diversity, i.e. chemodiversity, has been found in several plant species, of which some are of major ecological or economic relevance. Moreover, even within an individual plant there is substantial chemodiversity among tissues and across seasons. This chemodiversity likely has pronounced ecological effects on plant mutualists and antagonists, associated foodwebs and, ultimately, biodiversity. Surprisingly, studies on interactions between plants and their herbivores or pollinators often neglect plant chemistry as a level of diversity and phenotypic variation. The main aim of this Research Unit (RU) is to understand the emergence and maintenance of intraspecific chemodiversity in plants. We address the following central questions:
1) How does plant chemodiversity vary across levels, i.e., within individuals, among individuals within populations, and among populations?
2) What are the ecological consequences of intraspecific plant chemodiversity?
3) How is plant chemodiversity genetically determined and maintained?
By combining field and laboratory studies with metabolomics, transcriptomics, genetic tools, statistical data analysis and modelling, we aim to understand causes and consequences of plant chemodiversity and elucidate its impacts on the interactions of plants with their biotic environment. Furthermore, we want to identify general principles, which hold across different species, and develop meaningful measures to describe the fascinating diversity of defence chemicals in plants. These tasks require integrated scientific collaboration of experts in experimental and theoretical ecology, including chemical and molecular ecology, (bio)chemistry and evolution.
Keywords
Chemodiversity, biodiversity, plant-herbivore interactions, metabolomics, transcriptomics
State of the art and preliminary work
State of the art
Phenotypic differences among individuals within a population provide the raw material for natural selection and, when genetically based, lead to adaptive evolution. The importance of individual phenotypic differences is now recognised in various research fields (
- to characterise intraspecific plant chemodiversity at different levels,
- to study the ecological consequences of such diversity and
- to determine, how intraspecific plant chemodiversity is genetically determined and maintained.
The importance of variation in chemical phenotypes of organisms for shaping species interactions has often been neglected, although chemical communication is the most ancient mediator of information exchange among individuals. Specialised metabolites play an important role in the communication between plants and their environment. They serve as defences against herbivores or pathogens but also as attractants for beneficial organisms such as pollinators (
Differences in metabolite composition do not only occur among but also within individual plants (
Chemodiversity may be enhanced in response to the abiotic and biotic environment, in dependence of phenotypic plasticity (
Most theories on optimal defence production assume that the production of (new) specialised metabolites comes with biochemical costs. These costs are quite difficult to measure directly (
Traditionally, the most characteristic taxon-specific specialised metabolites are studied in detail (e.g., specific alkaloids in Solanaceae). More recently, large-scale untargeted metabolic fingerprinting and targeted profiling are used to gain a more complete picture of the overall chemodiversity in plants (
At the molecular level, metabolite diversification can be a result of various mechanisms, including gene duplication and neo-functionalisation, changes in transcriptional regulation or changes in enzymes involved in the biosynthesis of plant metabolites (
(Joint) preliminary work
Our RU brings together leading experts who have conducted (often jointly) preliminary work in all relevant research fields and are well experienced in their disciplines:
Sybille B. Unsicker (P1) is a chemical ecologist with special interest in the direct and induced defences of woody plant species against their natural enemies such as insect herbivores and pathogens (e.g.,
Jörg-Peter Schnitzler (P2) is a plant physiologist and biochemist, who is particularly interested in deciphering the metabolic vocabulary of plants and analysing biochemical mechanisms underlying biological functions of plant volatiles (e.g.,
Anke Steppuhn (P3) is a molecular ecologist, who investigates plant defence strategies against herbivore feeding and egg deposition. She aims to understand the strategies of plants used to fend off herbivores, costs and benefits involved in defence under different environmental conditions and how plants perceive herbivore attack (e.g.,
Nicole M. van Dam (P4) is a molecular ecologist, studying the chemical and molecular mechanisms underlying interactions between plants and their environment, especially herbivores and higher trophic levels. She has a long-standing expertise in the metabolite analysis of above- and belowground herbivore-induced defence responses (e.g.,
Caroline Müller (P5) is a chemical ecologist, who investigates the role of natural products involved in communication between organisms. She is particularly interested in the inter- and intraspecific variation in plant responses to environmental challenges, in tissue-specific analysis of metabolites with special focus on phloem sap and in the role of factors modulating plant-antagonist interactions (e.g.,
Elisabeth J. Eilers (P6) is an ecologist, who investigates chemo-ecological aspects of mutualistic and antagonistic plant-insect interactions (e.g.,
Wolfgang W. Weisser (P7) is an insect ecologist, who is interested in biodiversity and in multitrophic interactions at the population, community and ecosystem level. He studies interactions between plants, herbivorous insects and their natural enemies as well as mutualists, and investigates the involvement of plant volatiles in these interactions, using T. vulgare as model (
Andrea Bräutigam (P8) is a computational biologist with special interest in network analysis and modelling. Her research focusses on elucidating the molecular composition of complex traits and their evolution (e.g.,
Meike J. Wittmann (P9) is a theoretical biologist broadly interested in theoretical ecology and population genetics, and in particular in the interplay between ecological and evolutionary processes (e.g.,
Robert R. Junker (P10) is an evolutionary ecologist, who focuses on plant-animal and plant-bacteria interactions. He investigates how volatile compounds affect the diversity and interaction patterns of insects and bacteria and how these organisms alter the phenotype of plant species (e.g.,
Project-related publications
Principle investigators highlighted
Bräutigam A , Mullick T, Schliesky S, Weber APM (2011) Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C3 and C4 species. J Exp Bot 62: 3093-3102
Calf OW, Huber H, Peters JL, Weinhold A, van Dam NM (2018) Glycoalkaloid composition explains variation in slug resistance in Solanum dulcamara. Oecologia 187: 495-506.
Clancy MV, Zytynska SE, Moritz F, Witting M, Schmitt-Kopplin P, Weisser WW, Schnitzler JP (2018) Metabotype variation in a field population of tansy plants influences aphid host selection. Plant Cell Environ 41: 2791-2805
Clavijo McCormick A, Irmisch S, Reinecke A, Boeckler GA, Veit D, Reichelt M, Hansson BS, Gershenzon J, Köllner TG, Unsicker SB (2014) Herbivore-induced volatile emission in black poplar: regulation and role in attracting herbivore enemies. Plant Cell Environ 37: 1909-1923
Eilers EJ , Pauls G, Rillig MC, Hansson BS, Hilker M, Reinecke A (2015) Novel set-up for low disturbance sampling of volatile and non-volatile compounds from plant roots. J Chem Ecol 41: 253-266
Geuss D, Stelzer S, Lortzing T, Steppuhn A (2017) An ovicidal response of Solanum dulcamara to insect oviposition. Plant Cell Environ 40: 2663-2677
Jakobs R, Schweiger R, Müller C (2019) Aphid infestation leads to plant part-specific changes in phloem sap chemistry, which may indicate niche construction. New Phytol 221: 503-514
Junker RR, Kuppler J, Amo L, Blande J, Borges R, van Dam N, ...., Unsicker SB, et al. (2018) Covariation and phenotypic integration in chemical communication displays: biosynthetic constraints and eco-evolutionary implications. New Phytol 220: 739-749
Weisser WW , Roscher C, Meyer ST, Ebeling A, Luo GJ, Allan E, et al. (2017) Biodiversity effects on ecosystem functioning in a 15-year grassland experiment: Patterns, mechanisms, and open questions. Basic Appl Ecol 23: 1-73
Wittmann MJ, Fukami T (2018) Eco-evolutionary buffering: rapid evolution facilitates regional species coexistence despite local priority effects. Am Nat 191: E171-E184
Objectives, concept and approach
Common objectives of the RU
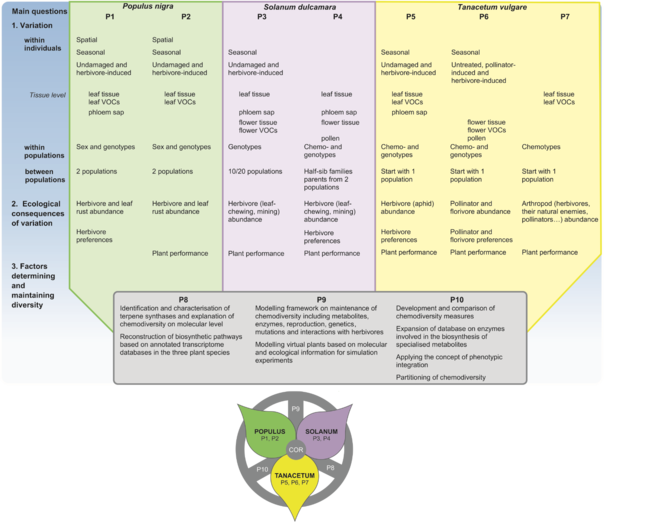
In the envisaged RU, we will investigate the ecology and evolution of intraspecific, including intra-individual, chemodiversity in three focal plant species. Combining field and laboratory studies with untargeted metabolomics and targeted metabolite profiling as well as genetic tools, statistical data mining and modelling, we want to elucidate the ecology and evolution of intraspecific chemodiversity of plants.
Our central questions are:
- How does plant chemodiversity vary across levels, i.e. within individuals, among individuals within populations, and among populations?
- What are the ecological consequences of intraspecific plant chemodiversity?
- How is plant chemodiversity genetically determined and maintained?
By addressing these questions, we aim to provide a scientific basis for understanding the role of intraspecific plant chemodiversity in natural plant communities, which may have important implications for pest control in crops, ecosystem functioning and nature restoration. We will take a combined experimental and theoretical approach, whereby ecological field and laboratory studies will be complemented with chemical and genetic analyses as well as mathematical modelling (Fig.
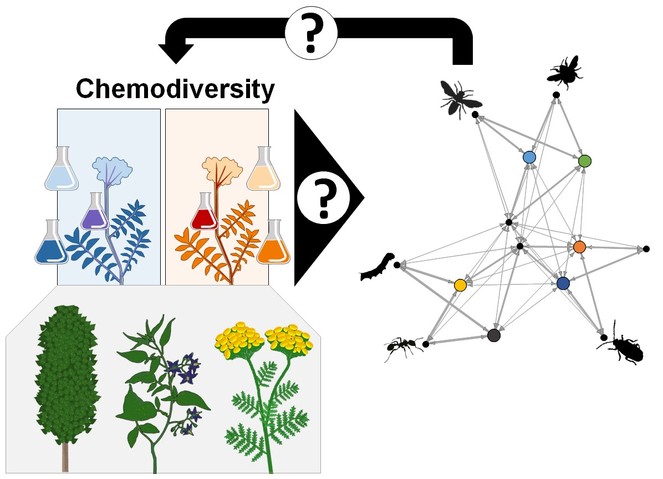
Conceptual framework of the proposed RU on the ecology and evolution of intraspecific plant chemodiversity. We will study chemical variation in different plant parts (flowers, nectar, pollen, leaves, phloem sap; roots will be included in a potential second funding period), among plant individuals within populations and among populations (left) as well as consequences on the plant-associated community (right) over space and time. The projects will focus on the tree Populus nigra and the herbs Solanum dulcamara and Tanacetum vulgare (lower panel, from left to right).
Within the first funding period, we aim to:
- develop a conceptual framework to describe chemodiversity, adopting and comparing common biodiversity measures for application to plant chemodiversity
- characterise the constitutive and induced plant chemodiversity within individuals, within populations and among populations with spatio-temporal resolution
- assess the effects of plant chemodiversity on herbivores with different feeding mode as well as pollinators and the contribution of phenotypic plasticity therein
- describe potential trade-offs between costs and benefits of plant chemodiversity on a community level
- identify and characterise genes coding for biosynthetic pathways relevant for plant-insect interactions and link chemodiversity to genetic features
- test whether patterns of chemodiversity in the three species support or contradict different hypotheses that aim to explain chemodiversity
- develop a new eco-evolutionary modelling framework for intraspecific plant chemodiversity
- generate specific data to parametrise the theoretical models
The overall aim of an envisaged second funding period is to implement the insights gained in the first phase into a broader context by:
- extending the geographic scale of study populations in all three species to comparatively asses the role of the breadth in environmental conditions accompanying their wide distribution
- including belowground intraspecific plant chemodiversity and effects on root herbivores
- investigating the causes and consequences of plant chemodiversity in more complex (natural) settings including microorganisms and higher trophic levels
- exploring the role of plant chemodiversity from the herbivore and pollinator perspective in more detail (e.g., in terms of counter-adaptations)
- developing genomic resources for the three study species such that chemodiversity can be linked to specific gene (and potentially epigenetic) variation and positioning in the genome
- incorporating more genetic realism into mathematical models for plant chemodiversity
Expected benefits of collaboration within the RU
This RU brings together a highly motivated, well-integrated and diverse group of scientists. We share a common vision on the ecology and evolution of plant chemodiversity. As a group, we can test hypotheses suggested by the currently available models explaining chemodiversity in parallel at multiple levels of complexity and synthesise the data to form well-supported conclusions. For example, the RU as a whole will be able to resolve if one of the key predictions from the screening hypothesis, i.e. that “plants maximise chemodiversity”, applies in the three study species. The combined expertise in this RU brings us in the unique position to thoroughly test this hypothesis in three distinct natural plant systems.
At the same time, each member has a unique area of expertise to generate the necessary synergies for a successful implementation of this joint project. Together, the competence of the applicants covers all methods necessary to address the questions and hypotheses of the individual projects as well as the data synthesis. Integrating field studies and classical ecological experiments with –omics techniques, bioinformatics and theoretical modelling will foster the exchange of knowledge and skills among RU members. Effective knowledge exchange will be ensured by regular meetings of all RU members and by lab visits of the young investigators (PhD students, postdoc). By these means, the young investigators will have access to a multifaceted method toolbox and expertise, which individual supervisors cannot offer.
For example, bioinformatic analyses of molecular data will be particularly supported by Bräutigam, who is an expert in transcriptome analyses and supervised machine learning, to study complex traits along evolutionary trajectories and to characterise traits at the molecular level (
Various metabolomics approaches have been developed in several labs involved in the RU. For instance, the collection of volatiles using push pull systems or passive trapping by polydimethylsiloxane (PDMS) tubes is well established by various PIs (
A challenge in the analysis of plant chemodiversity is the huge number of not yet chemically characterised metabolites. Given that concurrent identification and quantification of entire plant metabolomes is not possible to date, analytical tools such as mass difference networks (MDiN) and mass difference enrichment analysis (MDEA) can expand our capacity to analyse mass spectrometry data. These tools are established by Schnitzler (
Phloem sap collection by laser stylectomy or using ethylenediaminetetraacetate and analysis of the exudates is well established in the Müller group (
Investigating the role of intraspecific plant chemodiversity for herbivore resistance and its importance in shaping trade-offs among metabolites in affecting antagonists and mutualists is a common aim in this RU. Investigating this topic comprehensively will be enabled by the complementary expertise of different PIs on plant resistance mechanisms (
The ecology and evolution of intraspecific plant chemodiversity can only be understood by studying the consequences of this diversity in the field and in ecologically relevant experiments. Thus, a very important feature of the RU is to study the effects of chemodiversity in the field. Most PIs have extensive experience in studying interactions between plants and other organisms in common gardens and the field.
Finally, the integration of data (synthesis) is one of the key goals of the current RU. Combining the metabolic data from the different plant species and tissue types, we aim to identify patterns of co-variation in the composition of metabolites that can be aggregated in phenotypic integration values across an organisational hierarchy. Junker has been very successful in bringing together data from various researchers to study biosynthetic constraints and eco-evolutionary implications of the co-variation and phenotypic integration in chemical communication displays (
Expected key results in the short- and medium-term
We focus on three plant species differing in life-history traits to address the common key questions of this RU on the ecology and evolution intraspecific plant chemodiversity. The common approach used in this RU will strongly contribute to our understanding of how plant chemodiversity varies across different levels within selected plant species and how such chemodiversity affects interactions with other antagonistic and mutualistic organisms. We will determine how biotic factors influence the biosynthesis of specialised metabolites and reveal patterns of co-variation. In conclusion, we will gain novel insights on several levels of intraspecific plant chemodiversity and its functionality in interactions with other organisms.
Long-term goals
In the long-term, we aim to study additional layers of intra-individual differences. This will be achieved by including research on the chemodiversity of roots and interactions with belowground herbivores as well as with symbiotic or pathogenic microorganisms. Moreover, populations from a broader geographic range differing in selection histories will be included. We also intend to investigate the role of epigenetics in chemodiversity as a possibly important mechanism for the adaptation of perennial plants to varying herbivore pressures. Studying three plant species with different life-history traits in detail will allow us to refine models explaining evolutionary origins of chemodiversity.
Work programme including proposed research methods
For this RU on chemodiversity we propose to combine a variety of different experimental approaches with state-of-the-art analytical techniques and mathematical modelling approaches to characterise metabolic phenotypes with high resolution mass spectrometry. At the same time, we aim to develop a clearer definition of the term chemodiversity, whereby chemodiversity can be found at different hierarchical levels. This is needed to explicitly link metabolic patterns to community dynamics and biodiversity.
Research framework
Study systems: We focus our research on three study species that are ideal to address our research questions, P. nigra, S. dulcamara and T. vulgare. All three species exhibit a high degree of chemodiversity and naturally occur in Northern and Central European ecosystems including Germany. Although none of these species are classical model plants in molecular ecology, they fulfil essential characteristics for studying the eco-evolutionary relevance of chemodiversity also under natural selective regimes.
The three species selected for our projects have several features in common, which will allow us to transfer concepts and to determine more general principles. They are perennial and can be cloned easily, which is important for obtaining sufficient replicates and testing phenotypic plasticity while controlling for genetic background. Furthermore, all three species have been introduced to other continents. This widespread distribution will allow us in the long-term to compare populations with different selection histories on a large geographic scale. Most importantly, the three species have received attention based on their interesting metabolites, e.g. in the field of pharmaceutics (
Using a common experimental design, leaf volatiles will be measured in the three study species applying the same methodology [PDMS tubes (
Milestones and synergies
The expected benefits of collaboration (Expected benefits of collaboration within the RU), expected key results in the short- and medium-term (Expected key results in the short- and medium-term), and the long-term goals (Long-term goals), are outlined above and summarise our milestones and synergies. As an additional overarching milestone we need to reach a proper definition of ‘chemotype’ and ‘chemodiveristy’. As a working definition, we define a chemotype as a group of plants within a species, which can be distinguished according to the distinct and mostly heritable relative composition of compounds belonging to one prominent class of specialised metabolites, e.g., glycoalkaloids or terpenoids (
Moreover, for a synthesis of the various data from the individual projects, we aim to calculate the phenotypic integration of plant metabolites across an organisational hierarchy (mostly second year). The empirical data sets produced by the members of the RU in their individual projects (P1–P8) will provide the scientific basis to investigate the biosynthetic and genetic organisation of chemical trait co-variation and to evaluate the functional importance of phenotypic integration in plant metabolites (P10). In order to discriminate among functional, genetic, developmental and environmental causes of co-variation, phenotypic integration and modularity (
In conclusion, our combined approach will substantially contribute to our understanding of how plant chemodiversity varies across different levels, how such chemodiversity affects interactions with other antagonistic and mutualistic organisms and how it is maintained.
Research data and knowledge management
In addition to routine communication (by e-mail, phone and Skype), we will use the following means to ensure an efficient and stimulating exchange of knowledge:
Internal data sharing platform and data management: We have established a project share box in Sciebo (cloud storage platform of research institutions in North Rhine-Westfalia, guest status for members of the RU via Bielefeld University). This share box serves as the platform for data integration and management and for the exchange of method protocols within our group.
Public data repositories: We will create a detailed data management plan that will address all questions regarding data capture, storage, back-up, documentation, publication and long-term archiving of data collected at the beginning of this project. The Research Data Management Competence Center of Bielefeld University will assist the RU in developing procedures for Findable, Accessible, Interoperable, and Re-usable (FAIR) data during the project. Raw data of ecological and environmental parameters will be made available by publication with digital object identifiers (DOI). Modelling and simulation code will be placed under an open source licence. For both data and code, we will use repositories such as dryad or figshare, institutional repositories, or upload the material directly as supplementary information. Metabolomics and RNA-seq data will be uploaded to the corresponding repositories. MetaboLights is an open-access database for metabolomics experiments assigning studies a unique accession number as publication reference. Adhering to best practices in metabolomics data sharing will be done in collaboration with the de.NBI MASH project (Steffen Neumann, IPB Halle). Steffen Neumann offered his support to the RU. Prior to the upload of RNA-seq data to the European Nucleotide Archive (ENA) or gene expression omnibus (GEO), data will be stored in a network file system with automated back-ups (Bräutigam). Thus, all data and code connected to publications will be shared according to FAIR principles, available long-term and be quotable by DOIs.
Potential impact on the research area and local environment
All PIs involved in the RU are leading experts in their field and their home institutes have a high scientific reputation. Several researchers within the envisaged RU collaborate already successfully as evidenced by joint publications (see (Joint) preliminary work). Between and among researchers, the discussions leading to the RU have spawned collaborative efforts, which will strengthen over time, bringing the researchers and their institutions closer together and leading to fruitful collaboration. With this RU, we expect to become nationally and internationally visible as a scientifically strong consortium working on intraspecific plant chemodiversity. At the same time, we expect that our consortium will have a significant impact on the international community of chemical ecologists by building bridges among research disciplines, study organisms and study levels to achieve common overarching goals. This will happen when RU members present their work at international conferences, via publications in international journals including special issues on chemical diversity edited by the applicants, and by means of our workshops with national and international guests.
Measures to advance research careers
Early career researchers
The proposed RU includes several young scientists as PIs (Eilers, Steppuhn, Unsicker, Wittmann), who made already major contributions in their fields. Eilers joined the group of Müller in 2016 and is building up her independent research while pursuing a habilitation. Steppuhn was promoted to a W2 position (non-permanent) at Free University of Berlin in 2017. Unsicker is currently working on her habilitation. Wittmann started a position as Jun. Prof. (tenure-track) in 2017 at Bielefeld University. A successful participation in an internationally respected RU will increase the visibility of these young researchers, advance their careers and increase their chances to be tenured. Wittmann will be the Deputy Speaker of the RU, offering her the opportunity to gain experience in leading a large collaborative DFG-funded project.
Doctoral programmes
The young investigators employed in this RU will profit from structured doctoral programmes established at all involved universities, iDiv, Helmholtz and MPI-CE. Furthermore, they will choose a second mentor who is a PI in the RU but not their direct supervisor. The mentee will meet with her/his second mentor at least twice a year and will discuss the ongoing work, planning of experiments and manuscripts as well as career prospects. Financed by the travel budgets of the individual projects (P1–P10), the young investigators will be enabled to present their research to the scientific community and to network at national and international conferences.
Gender equality, career and family
Our PIs can serve as role models in various ways. Seven out of 10 (70 %) of our PIs are female and can serve as role models of successful female scientists. Several of our PIs (see CVs) can demonstrate to young scientists that it is possible to raise children and be successful in academia. When hiring young investigators for the projects, a balanced gender ratio will be strived for. Female junior scientists will be encouraged to participate in mentoring programmes and soft skill workshops adapted to the particular challenges of women in science such as movement (Bielefeld University) or Minerva-FemmeNet (MPG). All involved institutions have policies in place to ensure gender equality and family-friendly work conditions. Additionally, funds are requested for flexible childcare and home office supplies for all scientists with care tasks (for details see Project-specific Workshop Module). Furthermore, PIs will raise their awareness about the diversity of students (different genders, cultural backgrounds, etc). They will promote equality in chances and work against discrimination and under-representation by taking part in trainings offered by their institutions and staying informed via, e.g., https://www.working-between-cultures.com.
National and international cooperation and networking
The networking mechanisms described above (Expected benefits of collaboration within the RU) and the planned retreats and workshops (see below) will establish our RU as a national research consortium leading this field. National and international scientists will be invited to our workshops to stimulate discussions about chemodiversity in our scientific community and to encourage our young investigators to start building scientific networks. Furthermore, participation in international conferences such as the Gordon Research Conference on Plant-Herbivore Interaction as well as meetings with a focus on plant ecology and evolution will stimulate international cooperation and networking.
Coordination
Description of how joint objectives and the joint work programme will be implemented in the coordination project
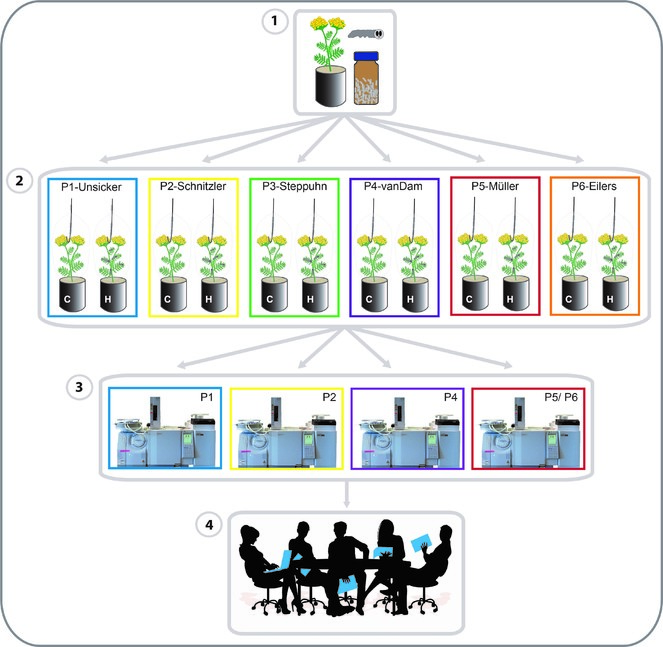
The coordination project and thus the speaker of the RU will facilitate the communication (including data management) and cooperation among the geographically dispersed groups and their research teams. The communication within the RU is based on regular meetings of all members but also on exchange between individual groups. For exchange between all groups and the scientific community, retreats and workshops will be coordinated as listed below. Exchange among individual groups will be mostly realised by mail, phone and skype or potentially video conferences. Webinars may enable virtual participation in seminars of general interest. For method standardisation, a ring trial for volatile collection is planned (see Coordination Module, Fig.
Anticipated total duration of the project
Three years.
Requested modules
Coordination Module
For administrative assistance as part of coordination, we apply for one service assistant (TVL E8, 50 %) for the speaker of the RU, Müller. The service assistant will provide the services for the administrative management of the RU, manage the central budget, administer the common website and shared data platform (Research data and knowledge management) as well as help in scheduling, organising and implementing the retreats and workshops.
Furthermore, a student assistant (10 h/week for 36 months) is requested who will be involved in the following three tasks:
- Structuring of data and metadata format for data to be collected by all groups and stored centrally; support in data upload and management (student with bioinformatics background)
- Support in preparation and cultivation of T. vulgare clones used in the projects P5–P8
- Coordination of ring trial (preparation of material, comparative data analysis):
At the beginning of the RU, we plan a ring trial of volatile collection by PDMS and analysis by thermodesorption (TD)-GC-MS. Clones of T. vulgare will be distributed by P5 and P6 to different groups (Fig.
Metabolomics platforms exist in several groups. The PIs of the individual projects have made individual agreements where they will run their analyses for an efficient division of resources and instrument time over the various labs. The PIs leading the platforms will be in regular exchange of expertise, methods and protocols. For ready comparison of metabolome data gathered by the different groups we will establish common sample preparation and measurement methods, as far as possible. Moreover, Schnitzler in collaboration with Philippe Schmitt-Kopplin (Helmholtz Zentrum München) established novel analytical methods such as mass difference networks, which can be applied on metabolomics and transcriptomics data sets. Schmitt-Kopplin will offer his expertise for data analysis in various projects.
RNA-seq analysis: An RNA-seq platform followed by single nucleotide variation (SNP)-based population analyses will be offered by Bräutigam. The platform includes the analytical set-up for quantitative transcriptome analyses of non-model species and the analytical set-up for transcriptome assembly, SNP discovery and quantification. We will offer a three-day RNAseq workshop ("Hackathon") to effectively transfer knowledge within the RU.
Advisory Board: We will set up an Advisory Board of three experts in the field with international reputation. These Board members will be invited to the workshops and may potentially host a young investigator for a lab visit and training in their lab.
Lab exchange of young investigators: The young investigators will be encouraged to visit other labs of PIs of the RU and beyond (e.g. potentially members of the advisory board) to be trained in methods that are important for their projects. For these visits, we apply for a lump sum of 15,000 €.
Network Funds Module
We aim to share metabolite standards and buy new standards, which will be distributed to all analytically working groups, to expand our databases needed for metabolite identification. To realise this goal we apply for a 10,000 € lump sum.
Gender Equality Measures in Research Networks Module
All participating institutions have explicit policies in place to ensure gender equality and family-friendly work conditions. These existing support structures and services will be strengthened by the following provisions:
- Flexible childcare and home office supplies for all scientists with care tasks
- Childcare during conferences or other events organised by the RU
- Flexible funds for, e.g., compensation for the absence of a RU member during pregnancy, care tasks for sick family members or parental leave
- Workshop on how to train a wide diversity of students and promote equal opportunities at all levels
- Specific communication trainings empowering young scientists to deal with implicit bias and micro-aggression, e.g. Active Bystander training by Scott Solder (http://www.scottsolder.com/)
For these measures, we apply for a lump sum of 15.000 € p.a.
Project-specific Workshop Module
Yearly retreats: All PIs and the young investigators involved in the projects will meet at least once per year for a 1-2 day retreat, which will be combined with a workshop. These retreats will be used to align the projects and to synthesise the most important findings and achievements of the RU. The young investigators will present their data to allow for feedback of other members of the RU and invited guest speakers. Guest speakers will give valuable input to the RU by reporting on adjacent topics. Furthermore, we will organise a “science speed dating” event where each young investigator talks for 5 min to each guest speaker (of course in the hope that longer conversations will follow) to improve networking skills and to get advice on academic careers. The young investigators will be encouraged to organise an own programme block directly before or after the retreat and workshop, in which they may exchange without their PIs and organise personal or scientific development courses depending on how they choose to allocate their time and financial support. For these young investigator modules, we apply for a lump sum of 5,000 € p.a. Having responsibility for the budget and the content will allow the young investigators to develop responsibility and deepen their scientific interactions while at the same time the self-developed programmes will address their actual needs.
Workshop 1: In the first year, we plan to organise a workshop on the topic “Plant Chemodiversity” to discuss the most useful chemodiversity measurements and indices developed by project P10 based on data provided by P1–P7. This workshop will be hosted by Junker in Marburg and international experts in biodiversity, chemical ecology and handling of complex metabolomic datasets will be invited.
Workshop 2: In the second year, the workshop will focus on “Evolution of Chemodiversity” and will be chaired by Wittmann. The workshop will take place in Leipzig at the German Centre for Integrative Biodiversity Research (iDiv) and co-organised by van Dam and members of her group. Guest speakers will be invited to discuss with us causes and consequences of chemodiversity.
International Workshop 3: A third workshop aims to discuss the findings of the RU in an international context. The “Synthesis” of our findings will be presented and discussed with international experts and we aim to write a joint paper on “The ecology and evolution of plant chemodiversity”. The workshop will be held at the Centre for Interdisciplinary Research (ZIF) of Bielefeld University and will be co-hosted by Eilers and Müller. At least 15 national and international experts in this field will be invited. To co-fund this workshop, we will also apply for funding at the ZIF.
Hackathon: In addition to the workshops, a “Hackathon” will be offered in the second year, which will be organised by Bräutigam, with input from Schnitzler and van Dam. All members of the RU will gather insight in transcriptome assembly, quantification and analyses for application in ecological and evolutionary concepts. The participants will meet in a conference room equipped with computers directly linked to the compute cluster at CeBiTec, Bielefeld University, and direct data access. Jointly, we will handle our data, share methods, scripts, programmes and ideas, pursue wild ideas and push our projects forward. This hackathon will foster interactions among the researchers which will strengthen the collaborations and give us room to try new and unusual approaches and share and develop new ideas.
Public Relations Module
A website will be established to advertise the general aims of our planned RU and to briefly introduce the aims of the individual projects potentially interesting to the scientific community as well as the public. The content will be regularly updated with current information, results, ideas and data from our projects. The estimated costs for this website will be around 3,000 €, to engage a professional to ensure a functional and attractive website that increases visibility. Dissemination of information to the group and the public will also make use of Twitter. Press releases will be issued on fundamental results via the press offices of the institutions.
In addition, we will train the PIs and especially the young investigators in science communication via the Public Engagement Training offered by City2Science (Herford, Germany). This training will be held in Bielefeld and will include sessions from “Science in Society” to “Responsible Research and Innovation” as well as sessions about the personal advantages and best practice examples of science-society dialogues. Furthermore, participants will design a dialogue-oriented communication idea for their research topic, which they then can present to the lay public at science fairs like the GENIALE in Bielefeld or the Long Night of Science in Leipzig and Jena. The costs for the training itself are 2,400 € plus an estimated cost of travel (200 € × 12 participants from outside of Bielefeld) and accommodation (80 € × 12 participants × 2 nights) of 4,320 € (Total: 6,720 €).
Funding program
Research Unit of Deutsche Forschungsgemeinschaft
Grant title
FOR3000
Conflicts of interest
The authors have declared that no competing interests exist.
References
-
Uncovering different parameters influencing florivory in a specialist herbivore.Ecological Entomology40(3):258‑268. https://doi.org/10.1111/een.12182
-
Comparison of the relative potential for epigenetic and genetic variation to contribute to trait stability.G3-Genes Genomes Genetics8(5):1733‑1746. https://doi.org/10.1534/g3.118.200127
-
Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis.New Phytologist196(2):596‑605. https://doi.org/10.1111/j.1469-8137.2012.04302.x
-
Linking aboveground and belowground interactions via induced plant defenses.Trends in Ecology & Evolution20:617‑624. https://doi.org/10.1016/j.tree.2005.08.006
-
Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses.Phytochemistry72(13):1497‑1509. https://doi.org/10.1016/j.phytochem.2011.01.038
-
Gypsy moth caterpillar feeding has only a marginal impact on phenolic compounds in old-growth black poplar.Journal of Chemical Ecology39(10):1301‑1312. https://doi.org/10.1007/s10886-013-0350-8
-
An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species.Plant Physiology155(1):142‑156. https://doi.org/10.1104/pp.110.159442
-
An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species.Plant Physiology155(1):142‑156. https://doi.org/10.1104/pp.110.159442
-
Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species.Journal of Experimental Botany65(13):3579‑3593. https://doi.org/10.1093/jxb/eru100
-
Intraspecific chemical diversity among neighbouring plants correlates positively with plant size and herbivore load but negatively with herbivore damage.Ecology Letters20(1):87‑97. https://doi.org/10.1111/ele.12713
-
Glycoalkaloid composition explains variation in slug-resistance in Solanum dulcamara.Oecologia187(2):495‑506. https://doi.org/10.1007/s00442-018-4064-z
-
Plant defences mediate interactions between herbivory and the direct foliar uptake of atmospheric reactive nitrogen.Nature Communications9:4743. https://doi.org/10.1038/s41467-018-07134-9
-
Costs of resistance in plants: from theory to evidence.Annual Plant Reviews47:263‑307. https://doi.org/10.1002/9781119312994.apr0512
-
Chemotypic variation in terpenes emitted from storage pools influences early aphid colonisation on tansy.Scientific Reports6:38087. https://doi.org/10.1038/srep38087
-
Metabotype variation in a field population of tansy plants influences aphid host selection.Plant Cell and Environment41(12):2791‑2805. https://doi.org/10.1111/pce.13407
-
Herbivore-induced volatile emission in black poplar: regulation and role in attracting herbivore enemies.Plant Cell and Environment37(8):1909‑1923. https://doi.org/10.1111/pce.12287
-
Plant genotypic diversity predicts community structure and governs an ecosystem process.Science313(5789):966‑968. https://doi.org/10.1126/science.1128326
-
The performance of polyclonal stands in short-rotation coppice willow for energy production.Biomass & Bioenergy8(1):1‑5. https://doi.org/10.1016/0961-9534(94)00077-7
-
What's the 'buzz' about? The ecology and evolutionary significance of buzz-pollination.Current Opinion in Plant Biology16(4):429‑435. https://doi.org/10.1016/j.pbi.2013.05.02
-
The ecological importance of intraspecific variation.Nature Ecology & Evolution2(1):57‑64. https://doi.org/10.1038/s41559-017-0402-5
-
Taschenlexikon der Pflanzen Deutschlands und angrenzender Länder: Die wichtigsten mitteleuropäischen Arten im Portrait.Quelle & Meyer
-
Butterflies and plants - A study in coevolution.Evolution18(4):586‑608. https://doi.org/10.2307/2406212
-
Solanaceae and Convolvulaceae: Secondary metabolites. Biosynthesis, chemotaxonomy, biological and economic significance.Springer-Verlag,Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-74541-9
-
Novel set-up for low-disturbance sampling of volatile and non-volatile compounds from plant roots.Journal of Chemical Ecology41(3):253‑266. https://doi.org/10.1007/s10886-015-0559-9
-
Natural products - a simple model to explain chemical diversity.Natural Product Reports20(4):382‑391. https://doi.org/10.1039/b208815k
-
The raison d'Être of secondary plant substances.Science129(3361):1466‑1470. https://doi.org/10.1126/science.129.3361.1466
-
Metabolic costs of terpenoid accumulation in higher plants.Journal of Chemical Ecology20(6):1281‑1328. https://doi.org/10.1007/BF02059810
-
Temporal changes affect plant chemistry and tritrophic interactions.Basic and Applied Ecology8(5):421‑433. https://doi.org/10.1016/j.baae.2006.09.005
-
Tritrophic interactions in wild and cultivated brassicaceous plant species.Wageningen University,Wageningen, NL.
-
Root herbivores and detritivores shape the aboveground multitrophic assemblage through plant mediated effects.Journal of Animal Ecology79:923‑931. https://doi.org/10.1111/j.1365-2656.2010.01681.x
-
Insect attraction to herbivore-induced beech volatiles under different forest management regimes.Oecologia176(2):569‑580. https://doi.org/10.1007/s00442-014-3025-4
-
Bioprofiling of Salicaceae bud extracts through high-performance thin-layer chromatography hyphenated to biochemical, microbiological and chemical detections.Journal of Chromatography A1490:201‑211. https://doi.org/10.1016/j.chroma.2017.02.019
-
A framework for predicting intraspecific variation in plant defense.Trends in Ecology and Evolution31(8):646‑656. https://doi.org/10.1016/j.tree.2016.05.007
-
From waste products to ecochemicals: Fifty years research of plant secondary metabolism.Phytochemistry68(22-24):2831‑2846. https://doi.org/10.1016/j.phytochem.2007.09.017
-
Pollination requirements of almond (Prunus dulcis): combining laboratory and field experiments.Journal of Economic Entomology111(3):1006‑1013. https://doi.org/10.1093/jee/toy053
-
Herbivore-induced changes in flower scent and morphology affect the structure of flower-visitor networks but not plant reproduction.Oikos125(9):1241‑1249. https://doi.org/10.1111/oik.02988
-
Two herbivore-induced cytochrome P450 enzymes CYP79D6 and CYP79D7 catalyze the formation of volatile aldoximes involved in poplar defense.Plant Cell25(11):4737‑4754. https://doi.org/10.1105/tpc.113.118265
-
Effects of intraspecific and intra-individual differences in plant quality on preference and performance of monophagous aphid species.Oecologia186:173‑184. https://doi.org/10.1007/s00442-017-3998-x
-
Volatile, stored and phloem exudate-located compounds represent different appearance levels affecting aphid niche choice.Phytochemistry159:1‑10. https://doi.org/10.1016/j.phytochem.2018.11.018
-
Aphid infestation leads to plant part-specific changes in phloem sap chemistry, which may indicate niche construction.New Phytologist221:503‑514. https://doi.org/10.1111/nph.15335
-
Metabolomic analysis of the interaction between plants and herbivores.Metabolomics5(1):150‑161. https://doi.org/10.1007/s11306-008-0124-4
-
Floral scents repel facultative flower visitors, but attract obligate ones.Annals of Botany105(5):777‑782. https://doi.org/10.1093/aob/mcq045
-
Floral odor bouquet loses its ant repellent properties after inhibition of terpene biosynthesis.Journal of Chemical Ecology37:1323‑1331. https://doi.org/10.1007/s10886-011-0043-0
-
Covariation and phenotypic integration in chemical communication displays: biosynthetic constraints and eco-evolutionary implications.New Phytologist220:739‑749. https://doi.org/10.1111/nph.14505
-
UV-B mediated metabolic rearrangements in poplar revealed by non-targeted metabolomics.Plant Cell and Environment38(5):892‑904. https://doi.org/10.1111/pce.12348
-
Mycorrhiza-triggered transcriptomic and metabolomic networks impinge on herbivore fitness.Plant Physiology176(4):2639‑2656. https://doi.org/10.1104/pp.17.01810
-
A robust, simple, high-throughput technique for time-resolved plant volatile analysis in field experiments.Plant Journal78(6):1060‑1072. https://doi.org/10.1111/tpj.12523
-
Variation in volatile compounds from tansy (Tanacetum vulgare L.) related to genetic and morphological differences of genotypes.Biochemical Systematics and Ecology29(3):267‑285. https://doi.org/10.1016/S0305-1978(00)00056-9
-
Plants suppress their emission of volatiles when growing with conspecifics.Journal of Chemical Ecology39(4):537‑545. https://doi.org/10.1007/s10886-013-0275-2
-
Plant volatile emission depends on the species composition of the neighboring plant community.BMC Plant Biology19(58). https://doi.org/10.1186/s12870-018-1541-9
-
Drought stress and leaf herbivory affect root terpenoid concentrations and growth of Tanacetum vulgare.Journal of Chemical Ecology40:1115‑1125. https://doi.org/10.1007/s10886-014-0505-2
-
Morphological integration and developmental modularity.Annual Review of Ecology Evolution and Systematics39:115‑132. https://doi.org/10.1146/annurev.ecolsys.37.091305.110054
-
The relative importance of plant intraspecific diversity in structuring arthropod communities: A meta-analysis.Functional Ecology32(7):1704‑1717. https://doi.org/10.1111/1365-2435.13062
-
UV-B impact on aphid performance mediated by plant quality and plant changes induced by aphids.Plant Biology12(4):676‑684. https://doi.org/10.1111/j.1438-8677.2009.00257.x
-
Biological activity of alkaloids from Solanum dulcamara L.Natural Product Research23(8):719‑723. https://doi.org/10.1080/14786410802267692
-
Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46's role in a nicotine-mediated antipredator herbivore defense.Proceedings of the National Academy of Sciences of the United States of America111(9):1245‑1252. https://doi.org/10.1073/pnas.1314848111
-
Time-invariant differences between plant individuals in interactions with arthropods correlate with intraspecific variation in plant phenology, morphology and floral scent.New Phytologist210(4):1357‑1368. https://doi.org/10.1111/nph.13858
-
Crosstalk between above- and belowground herbivores is mediated by minute metabolic responses of the host Arabidopsis thaliana.Journal of Experimental Botany63(17):6199‑6210. https://doi.org/10.1093/jxb/ers274
-
Phytochemical variation in treetops - causes and consequences for tree-insect-herbivore interactions.Oecologia187:377‑388. https://doi.org/10.1007/s00442-018-4087-5
-
A chemical polymorphism in a multitrophic setting: Thyme monoterpene composition and food web structure.American Naturalist166(4):517‑529. https://doi.org/10.1086/444438
-
Jasmonate signalling in plants shapes plant-insect interaction ecology.Current Opinion in Insect Science14:32‑39. https://doi.org/10.1016/j.cois.2016.01.002
-
Extrafloral nectar secretion from wounds of Solanum dulcamara.Nature Plants2(5). https://doi.org/10.1038/nplants.2016.56
-
Transcriptomic responses of Solanum dulcamara to natural and simulated herbivory.Molecular Ecology Resources17(6). https://doi.org/10.1111/1755-0998.12687
-
Can optimal defence theory be used to predict the distribution of plant chemical defences?Journal of Ecology98(5):985‑992. https://doi.org/10.1111/j.1365-2745.2010.01693.x
-
Tansy.Weed Technology6(1):242‑244. https://doi.org/10.1017/S0890037X00034643
-
Explaining intraspecific diversity in plant secondary metabolites in an ecological context.New Phytologist201(3):733‑750. https://doi.org/10.1111/nph.12526
-
Characterization of poplar metabotypes via mass difference enrichment analysis.Plant, Cell and Environment40(7):1057‑1073. https://doi.org/10.1111/pce.12878
-
Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories.Ecology and Evolution4(18):3514‑3524. https://doi.org/10.1002/ece3.1155
-
Resistance at the Plant Cuticle. In: Schaller A (Ed.)Induced Plant Resistance to Herbivory.Springer,Netherlands,22pp.
-
From plants to herbivores: novel insights into the ecological and evolutionary consequences of plant variation.Oecologia187(2):357‑360. https://doi.org/10.1007/s00442-018-4126-2
-
Tree genetic diversity increases arthropod diversity in willow short rotation coppice.Biomass & Bioenergy108:338‑344. https://doi.org/10.1016/j.biombioe.2017.12.001
-
Eco-metabolomics and metabolic modeling: making the leap from model systems in the lab to native populations in the field.Frontiers in Plant Science9: ‑1556https://doi.org/10.3389/fpls.2018.01556
-
Drought and flooding have distinct effects on herbivore-induced responses and resistance in Solanum dulcamara.Plant Cell and Environment39(7):1485‑1499. https://doi.org/10.1111/pce.12708
-
Interactive responses of Solanum dulcamara to drought and insect feeding are herbivore species-specific.International Journal of Molecular Sciences19(12). https://doi.org/10.3390/ijms19123845
-
Gene duplications and the time thereafter - examples from plant secondary metabolism.Plant Biology12(4):570‑577. https://doi.org/10.1111/j.1438-8677.2009.00317.x
-
Risk of herbivore attack and heritability of ontogenetic trajectories in plant defense.Oecologia187:413‑426. https://doi.org/10.1007/s00442-018-4077-7
-
Current challenges in eco-metabolomics.International Journal of Molecular Sciences19(5):1385. https://doi.org/10.3390/ijms19051385
-
The transcriptional landscape of polyploid wheat.Science361(6403):eaar6089. https://doi.org/10.1126/science.aar6089
-
Multiple feeding stimulants in Sinapis alba host plants for the oligophagous leaf beetle Phaedon cochleariae.Chemoecology18:19‑27. https://doi.org/10.1007/s00049-007-0389-5
-
Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate.Proceedings of the National Academy of Sciences of the United States of America106(26):10865‑10870. https://doi.org/10.1073/pnas.0904113106
-
Insect-plant biology.2.Oxford University Press,Oxford, UK.
-
Inbreeding diminishes herbivore-induced metabolic responses in native and invasive plant populations.Journal of Ecology107:923‑936. https://doi.org/10.1111/1365-2745.13068
-
Arbuscular mycorrhiza-induced shifts in foliar metabolism and photosynthesis mirror the developmental stage of the symbiosis and are only partly driven by improved phosphate uptake.Molecular Plant-Microbe Interactions27(12):1403‑1412. https://doi.org/10.1094/MPMI-05-14-0126-R
-
High specificity in plant metabolic responses to arbuscular mycorrhiza.Nature Communications5https://doi.org/10.1038/ncomms4886
-
Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types.Plant, Cell & Environment37:1574‑1585. https://doi.org/10.1111/pce.12257
-
Habitat variation, mutualism and predation shape the spatio-temporal dynamics of tansy aphids.Ecological Entomology42:389‑401. https://doi.org/10.1111/een.12396
-
Why are defensive toxins so variable? An evolutionary perspective.Biological Reviews87(4):874‑884. https://doi.org/10.1111/j.1469-185X.2012.00228.x
-
Resistance management in a native plant: nicotine prevents herbivores from compensating for plant protease inhibitors.Ecology Letters10(6):499‑511. https://doi.org/10.1111/j.1461-0248.2007.01045.x
-
Silencing jasmonate signalling and jasmonate-mediated defences reveals different survival strategies between two Nicotiana attenuata accessions.Molecular Ecology17(16):3717‑3732. https://doi.org/10.1111/j.1365-294X.2008.03862.x
-
Heavy metal (hyper)accumulation in leaves of Arabidopsis halleri is accompanied by a reduced performance of herbivores and shifts in leaf glucosinolate and element concentrations.Environmental and Experimental Botany133:78‑86. https://doi.org/10.1016/j.envexpbot.2016.10.003
-
Both heavy metal-amendment of soil and aphid-infestation increase Cd and Zn concentrations in phloem exudates of a metal-hyperaccumulating plant.Phytochemistry139:109‑117. https://doi.org/10.1016/j.phytochem.2017.04.010
-
Intracontinental plant invader shows matching genetic and chemical profiles and might benefit from high defence variation within populations.Journal of Ecology106:714‑726. https://doi.org/10.1111/1365-2745.12869
-
Specificity of induction responses in Sinapis alba and their effects on a specialist herbivore.Journal of Chemical Ecology33(8):1582‑1597. https://doi.org/10.1007/s10886-007-9322-1
-
Root and shoot glucosinolate allocation patterns follow optimal defence allocation theory.Journal of Ecology105(5):1256‑1266. https://doi.org/10.1111/1365-2745.12793
-
Invertebrate herbivory along a gradient of plant species diversity in extensively managed grasslands.Oecologia150(2):233‑246. https://doi.org/10.1007/s00442-006-0511-3
-
Protective perfumes: the role of vegetative volatiles in plant defense against herbivores.Current Opinion in Plant Biology12(4):479‑485. https://doi.org/10.1016/j.pbi.2009.04.001
-
Competition mediates costs of jasmonate-induced defences, nitrogen acquisition and transgenerational plasticity in Nicotiana attenuata.Functional Ecology15:406‑415. https://doi.org/10.1046/j.1365-2435.2001.00533.x
-
Belowground herbivory and plant defenses.Annual Review of Ecology Evolution and Systematics40:373‑391. https://doi.org/10.1146/annurev.ecolsys.110308.120314
-
Identification of biologically relevant compounds in aboveground and belowground induced volatile blends.Journal of Chemical Ecology36(9):1006‑1016. https://doi.org/10.1007/s10886-010-9844-9
-
A heritable glucosinolate polymorphism within natural populations of Barbarea vulgaris.Phytochemistry67(12):1214‑1223. https://doi.org/10.1016/j.phytochem.2006.04.021
-
Pollen competition between morphs in a pollen-color dimorphic herb and the loss of phenotypic polymorphism within populations.Evolution72(4):785‑797. https://doi.org/10.1111/evo.13445
-
Ecological and genetic effects of introduced species on their native competitors.Theoretical Population Biology84:25‑35. https://doi.org/10.1016/j.tpb.2012.11.003
-
Seasonally fluctuating selection can maintain polymorphism at many loci via segregation lift.Proceedings of the National Academy of Sciences of the United States of America114(46):E9932‑E9941. https://doi.org/10.1073/pnas.1702994114
-
Eco-evolutionary buffering: rapid evolution facilitates regional species coexistence despite local priority effects.The American Naturalist191:E9932‑E9941. https://doi.org/10.1086/697187
-
High chemical diversity of a plant species is accompanied by increased chemical defence in invasive populations.Biological Invasions13:2091. https://doi.org/10.1007/s10530-011-0028-5
-
Genetic and chemical variation of Tanacetum vulgare in plants of native and invasive origin.Biological Control61https://doi.org/10.1016/j.biocontrol.2012.01.009
-
An herbivore-induced plant volatile reduces parasitoid attraction by changing the smell of caterpillars.Science Advances4(5):eaar4767. https://doi.org/10.1126/sciadv.aar4767