|
Research Ideas and Outcomes : Research Idea
|
|
Corresponding author: Carlos G Mingorance Gámez (nirnel@hotmail.com)
Received: 31 Aug 2018 | Published: 26 Sep 2018
© 2018 Carlos Mingorance Gámez
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Mingorance Gámez C (2018) Epigenetic function of Granulocytic nuclei? Designing a new line of research. Research Ideas and Outcomes 4: e29438. https://doi.org/10.3897/rio.4.e29438
|
|
Abstract
Granulocytes share the common feature of having a lobulated nucleus, a fact whose function has yet to be discussed in depth. This hypothesis suggests that the division of the nuclei follows an epigenetic purpose, separating genes into compartments with different regulatory mechanisms, which may be due to intrinsic factors like regulatory RNA or extrinsic factors like proteins.
This paper describes the outlines of a line of research for both the initial testing to test the hypothesis and the following descriptive studies, including potential clinical uses in the prognosis or diagnosis of diseases that present dysregulation of the number of lobes. The chosen approach is to study the pattern of distribution (random or otherwise) of genes amongst the lobes.
Keywords
Basophils, Eosinophils, Epigenetics, Fluorescent In-situ Hybridisation, Genetics, Haematology, Immunology, Neutrophils
Overview and background
It has long been a widely known fact that neutrophils, basophils and eosinophils have lobulated nuclei. Curiously, there seems to be an acceptance of this fact at face value and seldom has anyone raised the question of why they are that way.
When the shape of a biological structure has a constant set of characteristics, it is certain to have a reason. A biological property can be secondary to another phenomenon, but it is more common to have been conserved because it confers an evolutionary advantage. This is the approach this paper is going to follow.
Shape being a mostly mechanical property, one would ponder first whether it would serve a mechanical goal. Indeed, especially in the case of neutrophils, there is much more space for granules in the cytoplasm, but a purely mechanical goal does not explain why there is a set number of lobes in basophils and eosinophils (2 lobes) and neutrophils (3-4 lobes) and not a more variable number.
In some pathological conditions such as lack of B12 vitamin and leukemia, to name but two, there is a variation of the number of nuclei (
Objectives
The working hypothesis of this essay is that, although an additional mechanical function may not be discarded, granulocyte nuclei segmentation has an epigenetic function, dividing groups of genes amongst different lobes with different properties. The idea of nuclear topography having an impact on epigenetics is not a novel one, with different studies finding an association between nuclear regions and epigenetics in Plasmodium (
Since there are currently no other publications concerning this particular field, this article will try to show a research line as complete as possible, starting with the testing of the initial hypothesis. Next follows a descriptive study and more in depth research looking for the underlying mechanisms and possible clinical applications.
Implementation
If this hypothesis is true, there will be a pattern showing a consistent distribution of the genetic material amongst the lobes, as opposed to an even and random distribution pattern in which all distributions would have an equal chance to appear — even if, through the microscope, proportions appear different (Figs
If the pattern of distribution of genes between lobes is random, every possibility has an equal chance to happen — or a consistently different chance, if some lobes are more likely to contain a higher amount of genes due to their larger size or similar factors. On the other hand, if this hypothesis is true, the distribution pattern would not be random. Instead, both copies of the same gene would most likely be always together in the same lobe or always in different lobes. In addition, if the position of the lobes is relevant, not only would the copies always be together or separated, but they would also be in the same nuclear position. So we can find patterns with different percentages for the distributions of the loci (Figs
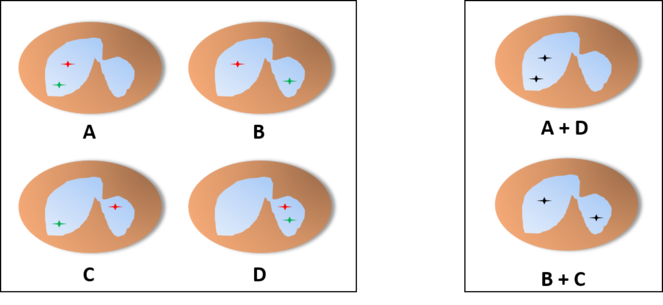
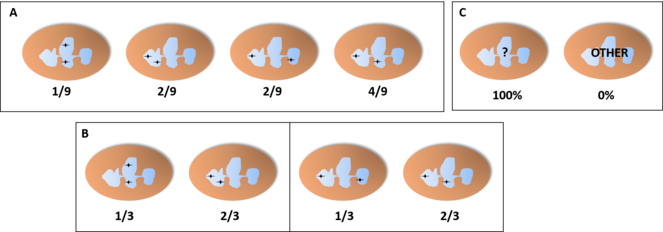
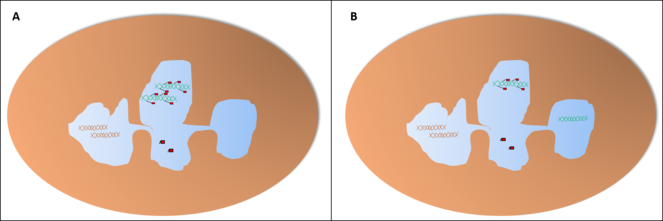
Probability of finding each distribution of the alleles in a basophil or eosinophil through the microscope, assuming an independent pattern (A, assuming an equal chance for both lobes), pattern of interdependent location independent of lobe (B) or pattern of interdependent location in a fixed lobe (C). In cases B and C, since these cells only have two lobes, we would find only cells with one of the distributions, so we would be unable to tell one of the patterns apart from the other.
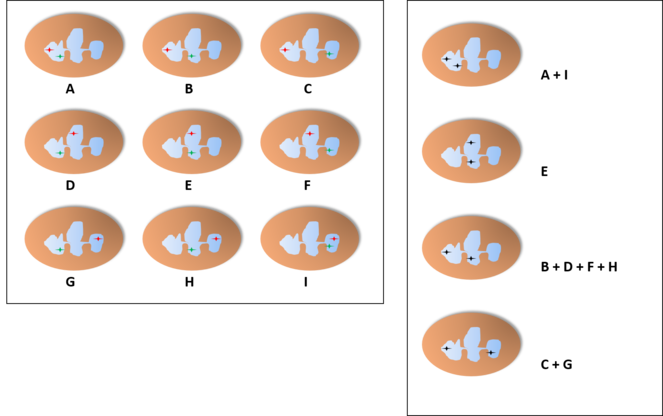
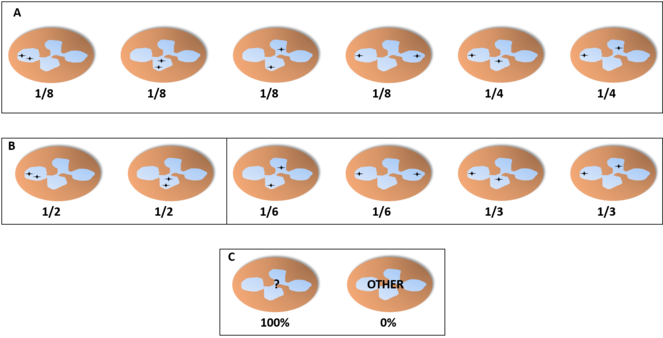
Probability of finding each distribution of the alleles in a neutrophil with three lobes through the microscope, assuming an independent pattern (A, assuming an equal chance for all lobes), pattern of interdependent location independent of lobe (B) or pattern of interdependent location in a fixed lobe (C). In B, we find two distributions with the shown proportions (either the two on the left or the two on the right) and the other two distributions have a probability of 0%. In C, one of the distributions would be present in 100% of the cells while the others would not appear.
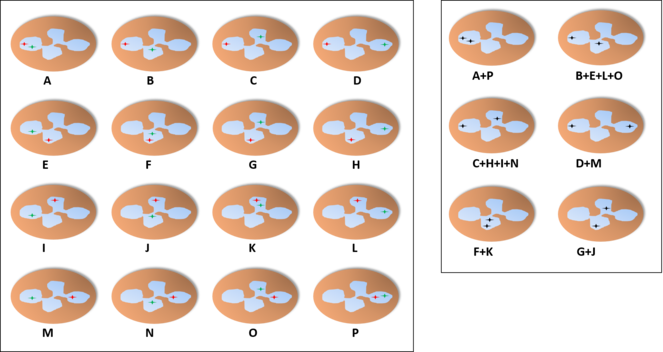
Probability of finding each distribution of the alleles in a neutrophil with four lobes through the microscope, assuming an independent pattern (A, assuming an equal chance for all lobes), pattern of interdependent location independent of lobe (B) or pattern of interdependent location in a fixed lobe (C). In B, we find two distributions with the shown proportions (either the two on the left or the four on the right) and the other distributions have a probability of 0%. In C, one of the distributions would be present in 100% of the cells while the others would not appear.
Three factors that could interfere with these percentages are:
- As mentioned, some lobes can have a different amount of genes because of their size or because of other circumstances, thus changing the proportions in the case of the random pattern.
- Certain genes might follow a random model while others are distributed following a pattern (so the test would be positive for some genes and negative for others).
- There might be different cell populations with their own patterns.
In the case of different probabilities because of varying lobe sizes, we could still recognise fixed proportions, according to which larger lobes would have a proportionally higher chance to contain the genes we are testing in the case of a random pattern — as opposed to a smaller lobe having a much higher affinity to the genes or even two lobes of widely different sizes with similar affinity to the genes, which would point to other explanations.
About the possibility of only some genes being positive, we must take into account that many genes, without the need for a fixed distribution, may be in linkage disequilibrium with genes distributed in a certain way. So, if this hypothesis is true, it would be extremely unlikely that a moderate number of randomly selected genes from different chromosomes would follow the exact theoretical random pattern, thus adding weight to negative results when studying more than one gene.
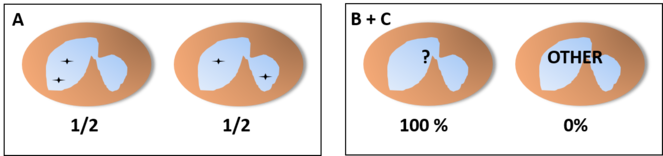
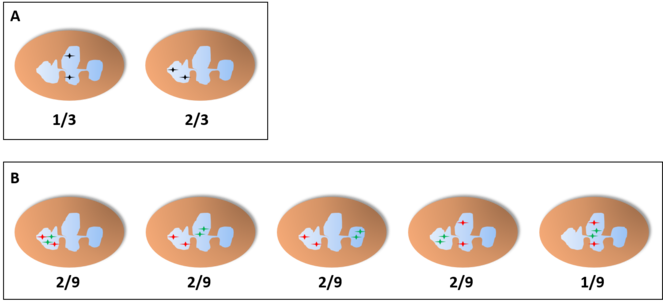
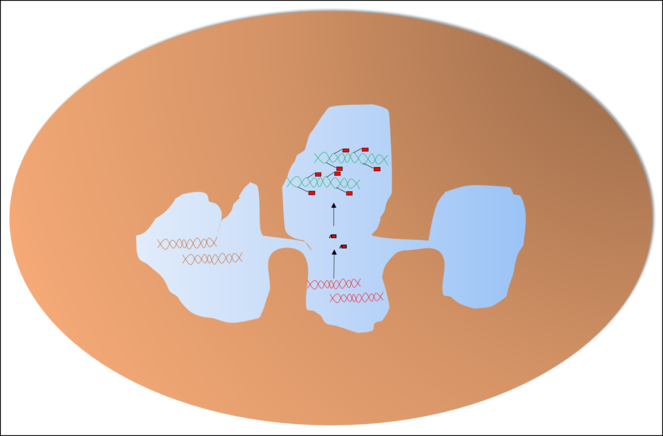
If we find different populations with their own patterns, it is unlikely that their proportions would be exactly like a random pattern, but even if that is the case, a joint study of different genes would help differentiate an independent pattern from an interdependent one (Fig.
Frequencies of joint random pattern studying a single target (A) or two (B) in a neutrophil with three lobes. Even if we find 2 populations of neutrophils with a pattern of joint distribution in a fixed lobe and showing the same frequencies and distributions shown in A, these two populations would show only two of the distributions shown in B with the same frequencies found before if the pattern is not actually independent of the lobe.
Technical issues
For the first stage of the experiment, preliminary testing the hypothesis, we would not need a large number of test subjects. A few blood samples from healthy volunteers would be enough to obtain a sufficient amount of neutrophils to use in a chi-square test. Only in the latter phases would we need a wider selection of subjects.
The ideal technique for the previously described experiment would be an in situ hybridisation combined with a nuclear membrane or background staining, but a very strict conservation of nuclear morphology is essential and current hybridisation protocols show a limited conservation of nuclear shape.
All of the experiments described in this paper need a way of combining an in situ hybridisation with nuclear staining and a practically perfect nuclear shape conservation. The deformation is most likely caused by fixation and more efficient fixation techniques can help solving the problem completely or partially (
The possible increase in nuclear loss of shape caused by denaturalisation temperatures could be avoided either by:
- Previously fixating the nuclear membrane in a way that protects it from thermal deformation.
- Achieving the separation of DNA strands by methods less invasive than high temperature.
- Hybridising with an intronic sequence of a highly transcribed hnRNA instead of DNA, thus eschewing the need to denaturalise the DNA at all.
The less invasive methods mentioned in point 2 might be genetic tools like target-specific transcription factors and other signals used by the cell to separate both strands. Depending on how the transcription of each locus is regulated, it could be less reliable than high temperature. If we want our probe to arrive at the vacant sequence and stay hybridising, we may also need to prevent the RNA polymerase from joining or transcribing the locus or to employ nucleic acids with higher affinity for DNA than the other DNA strand, such as peptide nucleic acid (PNA) or locked nucleic acid (LNA) (
Hybridisation with an intronic sequence of hnRNA, as mentioned in point 3, would solve the problem of having to separate both strands of DNA, but it would be extremely difficult to wash away the excess probe without depleting the nucleus, so we may need to employ FRET or hairpin probes that only emit their signal when they have reached the target.
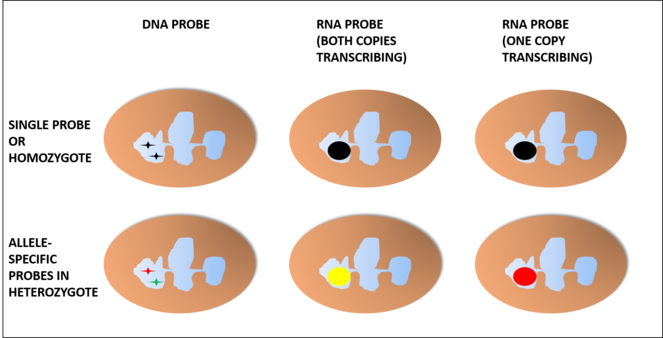
The main problem in this case would be telling apart two copies of the gene being transcribed in the same lobe from only a single copy being transcribed at all, since the probes would not join a single spot of the strand but rather a region of the nucleus with copies of the RNA strand (Fig.
When hybridising with intronic hnRNA, we would need an additional probe in order to discard false positives because of a copy of the gene not being transcribed and it would only work in heterozygotes. In this case, using red and green probes for different alleles would show as yellow if both copies are transcribing in the same lobe or as red or green if only one of them is being transcribed.
Underlying mechanisms
The reason different loci can be distributed between following a fixed pattern amongst the lobes may be that regulatory factors present in one lobe are absent in other.
Another option, in the case of neutrophils, is that some unused genes for this line (or the unused copy of a gene) are being processed in one lobe to convert them into neutrophil extracellular traps (NETs). DNA in these loci would be treated in such a way as to keep it prepared for quick release, but not available for gene transcription.
This said, the NET would not be enough to explain eosinophil and basophil segmentation if we take it on its own, so it would more likely be either a matter of gene regulation or both. At any rate, it may help to explain potential patterns in which both copies of a gene are present in different lobes in neutrophils (Fig.
About the mechanisms involved in either or both of these cases:
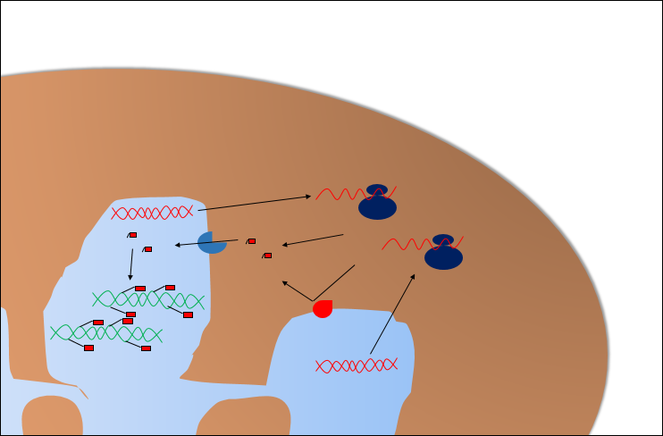
- It can be as simple as regulatory RNA acting only in the lobes that their genes are located. If this hypothesis is the only true one, then the physical barriers themselves are enough to achieve the desired goal (Fig.
10 ). - On the other hand, the process can be as complex as protein transcription factors or DNA-modifying proteins being distributed heterogeneously. In this case, since these factors are synthesised in the cytoplasm, the responsible mechanism would be differential migration, which would point to specific structures in or near nuclear pores amongst the lobes (Fig.
11 ).
Protein transcription factors would be synthesised in the cytoplasm, so their genes can be in any lobe. Their presence in each lobe would depend on the properties of its nuclear pores. In the image, red pore complexes would reject the factor while the blue ones facilitate their importation into the lobe.
Both of these mechanisms can be present at the same time, so experimentally proving one of them does not mean the other one is neither true nor false.
Descriptive works
If this theory is proven, the next step would be a descriptive study in which we take groups of loci to study at the same time, so we learn which genes are grouped together. The different lobes would receive names according to their properties or DNA composition. The nomenclature here proposed uses the Greek alphabet:
- Alpha for the lobe in which the centromere of chromosome 1 is (or the next chromosome in numeric order, if this centromere’s position is distributed at random).
- Beta for the lobe in which is the next centromere in numeric order that is not in lobe Alpha and is not randomly distributed.
- Gamma, delta and so on, following the stated criteria in cells with more than two lobes, including polysegmented cells.
A better, more descriptive nomenclature can be devised once we know more about the eventual specific properties and function of the different lobes.
In this stage, it would be desirable to obtain blood from a wider pool of healthy subjects so we can attest for individual and ethnic variation.
At this point in the research, since we would have proven the existence of a non-random pattern, it might be worth trying to apply chromosome painting instead of short probes, which may have appeared too expensive in the early steps but would be much more practical now.
Looking for the molecular mechanisms
After all of these experiments are done, assuming we find evidence of a non-random pattern of distribution of DNA, the last foreseeable step (which can be taken at the same time as the clinical approach) is to find out which mechanisms cause this phenomenon. Since the theories mentioned above are not mutually exclusive, each must be tested on its own.
Regulatory RNA can be easily tested, in theory. All we need to do is to detect their presence exclusively in the lobes along with their coding loci.
Protein transcription factors or DNA-modifying enzymes can be also studied by looking for them directly, but the pore complexes in different lobes should also be analysed.
In the case of NETosis, it can additionally be useful to search for DNA modifications in each lobe.
These seemingly easy tasks come with the added challenge of having to combine these techniques with at least one lobe identification. This longer-term problem will no doubt be more easily addressed, if needed and when it presents itself, due to the accrued experience we would have in this field at that point.
Impact
With the descriptive study finished in healthy subjects, we should next study the patterns of patients with polysegmented or hyposegmented cells, being on the watch for differential patterns that serve as a prognostic or diagnostic criterion in each of the studied diseases. In the long run, it could be extended to haematological diseases without segmentation issues, especially if we find more than one cell population with different patterns.
Uses in daily clinical practice
If the previous step shows a potential clinical usefulness, the most cost-effective technique for routine use would be using a probe for each involved locus (provided there are not technical issues that prevent us from employing this approach). Developing standard labelling for each lobe can make the work of probe designers simpler, be it through the addition of lobe-specific loci probes or through the detection of specific nuclear proteins, in the nuclear membrane or otherwise.
Conclusions
We are facing an obvious question that has not been raised before. Unfortunately, new experimental techniques are needed in order to properly study it but, should they be developed, we have a clear path for the investigation to follow.
Even though there is a clear possibility for this subject to have repercussions on some diseases, the first step would be to investigate it from a descriptive viewpoint before we can try a clinical approach, which might discourage some clinically orientated groups from investing in this line at first, but the potential here, especially for groups with less direct clinical implication, is large enough that others will surely try.
Hosting institution
Universidad de Granada, Spain
Ethics and security
Since the only interaction with test subjects would be during the extraction of a blood sample, the ethical concerns for this research are practically null and only an informed consent would be needed to proceed with any part of the research.
References
-
Applications of peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) in biosensor development.Analytical and Bioanalytical Chemistry402(10):3071‑3089. https://doi.org/10.1007/s00216-012-5742-z
-
Leukocyte Nucleus Segmentation and Nucleus Lobe Counting.BMC Bioinformatics11(1):558. https://doi.org/10.1186/1471-2105-11-558
-
A modified fluorescence in situ hybridization protocol for Plasmodium falciparum greatly improves nuclear architecture conservation.Molecular and Biochemical Parasitology173(1):48‑52. https://doi.org/10.1016/j.molbiopara.2010.04.006
-
Nuclear Compartmentalization Contributes to Stage-Specific Gene Expression Control in Trypanosoma cruzi.Frontiers in Cell and Developmental Biology5:8. https://doi.org/10.3389/fcell.2017.00008
-
Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum.International Journal for Parasitology40(1):109‑121. https://doi.org/10.1016/j.ijpara.2009.09.002