|
Research Ideas and Outcomes : Grant Proposal
|
|
Corresponding author: Roberto Toro (rto@pasteur.fr)
Received: 27 Mar 2018 | Published: 30 Mar 2018
© 2018 Roberto Toro, Rembrandt Bakker, Thierry Delzescaux, Alan Evans, Paul Tiesinga
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Toro R, Bakker R, Delzescaux T, Evans A, Tiesinga P (2018) FIIND: Ferret Interactive Integrated Neurodevelopment Atlas. Research Ideas and Outcomes 4: e25312. https://doi.org/10.3897/rio.4.e25312
|
|
Abstract
The first days after birth in ferrets provide a privileged view of the development of a complex mammalian brain. Unlike mice, ferrets develop a rich pattern of deep neocortical folds and cortico- cortical connections. Unlike humans and other primates, whose brains are well differentiated and folded at birth, ferrets are born with a very immature and completely smooth neocortex: folds, neocortical regionalisation and cortico-cortical connectivity develop in ferrets during the first postnatal days. After a period of fast neocortical expansion, during which brain volume increases by up to a factor of 4 in 2 weeks, the ferret brain reaches its adult volume at about 6 weeks of age. Ferrets could thus become a major animal model to investigate the neurobiological correlates of the phenomena observed in human neuroimaging. Many of these phenomena, such as the relationship between brain folding, cortico-cortical connectivity and neocortical regionalisation cannot be investigated in mice, but could be investigated in ferrets.
Our aim is to provide the research community with a detailed description of the development of a complex brain, necessary to better understand the nature of human neuroimaging data, create models of brain development, or analyse the relationship between multiple spatial scales. We have already started a project to constitute an open, collaborative atlas of ferret brain development, integrating multi-modal and multi-scale data. We have acquired data for 28 ferrets (4 animals per time point from P0 to adults), using high-resolution MRI and diffusion tensor imaging (DTI). We have developed an open-source pipeline to segment and produce – online – 3D reconstructions of brain MRI data.
We propose to process the brains of 16 of our specimens (from P0 to P16) using high-throughput 3D histology, staining for cytoarchitectonic landmarks, neuronal progenitors and neurogenesis. This would allow us to relate the MRI data that we have already acquired with multi-dimensional cell-scale information. Brains will be sectioned at 25 μm, stained, scanned at 0.25 μm of resolution, and processed for real-time multi-scale visualisation. We will extend our current web-platform to integrate an interactive multi-scale visualisation of the data. Using our combined expertise in computational neuroanatomy, multi-modal neuroimaging, neuroinformatics, and the development of inter-species atlases, we propose to build an open-source web platform to allow the collaborative, online, creation of atlases of the development of the ferret brain. The web platform will allow researchers to access and visualise interactively the MRI and histology data. It will also allow researchers to create collaborative, human curated, 3D segmentations of brain structures, as well as vectorial atlases. Our work will provide a first integrated atlas of ferret brain development, and the basis for an open platform for the creation of collaborative multi-modal, multi-scale, multi-species atlases.
Keywords
Neuroanatomy, Brain Development, Ferret, Open Science, Histology, Atlas
Relevance to the call
Objectives of the project
The first postnatal weeks in ferrets recapitulate many of the main stages in the construction of a complex mammalian brain. Unlike mice, ferrets develop a rich pattern of deep neocortical folds and cortico-cortical connections (
We propose to:
-
Process the brain of 16 of the specimens that we have already MRI scanned (P0-P16) using high-throughput 3D histology, staining for cytoarchitectonic landmarks, stem cells and neurogenesis,
-
Scan the complete series of histological brain slices at cell-scale resolution,
-
Process the data for real-time, multi-scale (deep-zoom) visualisation,
-
Make all the data acquired available online to the community through a web portal and HBP's Unified Portal,
-
Develop a collaborative tool to interactively annotate the data, segment brain regions and build brain atlases.
Each of these objectives will be described in more detail in section 2. Overall, we aim at providing the HBP and the research community with an integrated developmental atlas of the ferret brain, and to develop interactive online tools to facilitate and foster scientific collaboration.
Relevance to the topic
Our project is mainly relevant to area 4: Neuroinformatics, partnering with work package WP4.9 “Atlases for other species”, because it proposes the construction of atlases of ferret brain development, which is a species not covered by the HBP.
Our proposal also responds to the requirements of area 7: Medical informatics, work package WP7.12 “Methods and tools”, because we propose to develop a new type of open web tool for the collaborative, online analysis of MRI and histological data. We propose to use this tool for the visualisation and analysis of the ferret brain, but it will be equally suitable for data from mouse or human.
Finally, our proposal is also relevant to area 10: Ethics and Society, work-package WP10.8 “Public outreach”. The development of a web platform opens the possibility to launch crowdsourcing projects or citizen science projects, where the general public is directly engaged in the analysis of scientific data. This type of outreach has proven extremely successful in projects such as FoldIt (http://fold.it, a protein folding game) or EyeWire (http://eyewire.org, retina connectivity crowdsourcing), and we have already started a similar effort with our Brain Catalogue (http://braincatalogue.org, multi-species brain segmentation crowdsourcing).
Complementarity and potential synergies with the Flagship Core Project
The HBP focuses primarily on constructing two models: one for the mouse brain and one for the human brain. The step between the two is large, not only in terms of size – it represents a factor of about 1,000 (
The proposed project will be well integrated with the core of the HBP consortium. First, it will use the ICT infrastructure in the HBP to store the data and make it accessible. For this our team will interface with Martin Telefont who serves as the data integrator within the Neuroinformatics subproject. Specifically, the data will be incorporated in the knowledge space and the unified portal. Second, it will use the tools for image analysis that are developed for the Neuroinformatics Platform (NIP). For this we will interface with José María Peña, who leads the structural analysis work package, and serves as an advisor. Third, because it will add a new atlas to the NIP to augment the mouse and human, for which we will interface with Jan Bjaalie and Katrin Amunts, respectively who are responsible for these atlases.
The proposed studies connect to two research themes of the Dutch Roadmap ICT of the so called topsectoren. First, our goal is to develop a ferret brain atlas and facilitate other researchers with populating it with their data. This atlas and associated data will become available from the knowledge space and unified portal that is being developed within the HBP-core consortium, and from which the data will be made open and publicly accessible. Hence, the proposed studies contribute to the research theme “ICT for a connected world (Data interoperability)”. The data that will be collected consists of large stacks of high-resolution imaging data, for which data-mining techniques are developed and applied to extract for each brain area cell counts and to link data modalities across scales (DTI, MRI, gene-expression, anatomy) within a predictive modelling framework. As such the proposed studies also contribute to the research theme entitled “Data, data, data (Big Data)”.
Scientific and Technical Quality
State of the art and expected progress beyond state of the art
Our proposal comprises two main components:
- The acquisition and processing of multi-modal, multi-scale data from ferret brain development, the preparation of the data for sharing and visualisation, and
- The development of neuroinformatics tools for the collaborative construction of ferret brain atlases.
Building a new brain atlas is a long and arduous work. Detailed brain atlases are therefore available for only a handful of species, such as mice, rats or macaques. We lack brain atlases for many widely studied species, such as the ferret. Currently, researchers interested in ferret brain development rely on figures from papers, can access the few histological slices from a single adult specimen available at http://brainmuseum.org/Specimens/carnivora/ferret, or the labelled brain slices available at http://didier.theearlab.org/FerretBrainstemAtlas.html (intended to describe the organisation of the adult ferret brainstem). Concerning freely available data, the group of T2 MRIs that we have made available is, to the best of our knowledge, the only existing one (http://siphonophore.org/ferretproject, Fig.
Our proposal intends to improve the current situation by building a large, open, multi-modal, multi-scale dataset covering the first weeks of ferret brain development, with several specimens per time point. The resolution will be several orders of magnitude larger than in the existing projects, with sub- micrometer in-plane resolution, and up to 200 slices per staining modality, per individual. This will provide the community with a thorough description of ferret brain development, of quality comparable to those available for mice and macaques (Allen Institute), or human (BigBrain project). Additionally, the online platform that we have started to build should provide a way to create, collaboratively, atlases for species less popular than mice and rats.
Many tools are available today for online visualisation of histology data, MRI data and brain atlases: the Allen Institute for Brain Research platform (http://brain-map.org), the Mouse Brain Architecture project (http://brainarchitecture.org), the open source PivotViewer from the Computational Biology Research Group of the University of Oxford (http://www.cbrg.ox.ac.uk/data/pivotviewer), BrainBrowser from McGill University (http://brainbrowser.cbrain.mcgill.ca, Partner 4), the Scalable Atlas from INCF/Radboud University Nijmegen (http://scalablebrainatlas.incf.org, Partner 3, Fig.
The feature that lacks, however, in all current platforms is the ability to collaborate on the creation of an atlas: draw region borders, annotate them, and discuss/resolve conflicts between contributions. This aspect is present in the context of neuronal reconstruction in EyeWire from MIT (http://eyewire.org, closed source), and it will inspire us to extend the tools which we have built for brain segmentation (http://braincatalogue.org, Fig.
Scientific description of the project and research method
Overall research approach: The ferret as an animal model for human neuroimaging
The development of a human brain is the proliferation and migration of billions of cells (
Ferrets are recognised as an exceptional animal model to study brain development and may provide important information for the interpretation of human neuroimaging data (
Methodology and experimental protocols: An integrated, reliable, open-science approach
Our aim is to map ferret brain development by integrating multiple scales and imaging modalities. We have already acquired neuroimaging data (7 Tesla, high-resolution quantitative T2 MRI, DTI). Our methodology will be as follows:
- Process the whole brains (cerebrum + cerebellum) of 16 of the specimens that have already been MRI scanned (4 brains at P0, P4, P8 and P16) using high-throughput 3D histology (25 μm slice thickness), staining for cytoarchitectonic landmarks (Nissl and NeuN immunohistochemistry, IHC, stains), stem cells (Pax6 IHC staining) and neurogenesis (Tbr2 IHC stain). Brain sectioning and staining will be performed at Neuroscience Associates (http://www.neuroscienceassociates.com), with whom we have already worked in the past. Neuroscience Associates have developed a platform to section and stain several brains in parallel (MultiBrain technology). This ensures strictly controlled sectioning conditions and obtain a consistent staining quality for all specimens. During sectioning, high-resolution pictures of the block-face are taken, for later correction of artefacts during 3D reconstruction.
- Scan the complete series of brains for virtual microscopy. We will scan all the slices (N~3,200) at an in-plane resolution of 0.25 μm (20x magnification) using an AxioScanZ.1 Zeiss virtual microscopy scanner at the Institut Pasteur, capable of scanning a field of view corresponding to 50x16 brain slices (double glass slides). Data will be stored in Carl Zeiss Imaging format (CZI format), which has been devised to handle large data files and meta-data, and is supported by several open source image-analysis software such as ImageJ and Fiji.
-
Process the data for multi-scale, deep-zoom visualisation (DZI format). This data format allows for a resolution-independent, real-time zoom-in/out of the images. During the project only the images in the sectioning plane (coronal) will be processed. However, the correction of sectioning artefacts should allow us posteriorly to reconstruct 3D histology volumes and process virtual sections for deep-zoom visualisation. We will benefit of the High Performance Computing platform of the Institut Pasteur to process and store the data, as well as HBP's neuroinformatics platform.
-
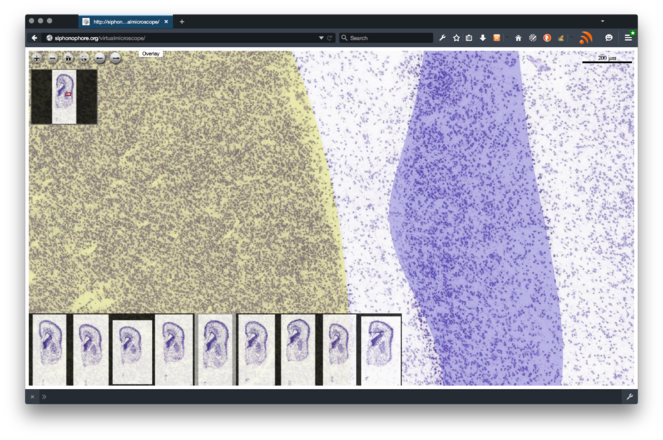
Make all the data acquired available to the community through an interactive web portal and HBP's Unified Portal. The interactive web portal will allow for real-time, online visualisation of the microscopy data. The data will be released under a Creative Commons licence, the application will be based on open-source technology (Open Seadragon, jQuery, etc.), and its code released open-source and made available as a tool within the HBP Neuroinformatics Platform. By making all code available to the community we will ensure a faster detection of bugs, as well as a distributed maintenance and extension of the platform. A first functional prototype of the web application is available at http://siphonophore.org/virtualmicroscope (see Fig.
4 ). To ensure reusability and interoperability, data will be also made available through HBP's Unified Portal. -
Develop, using the deep-zoom visualisation interface, a collaborative tool to manually annotatw the data, segment brain regions and build brain atlases. This tool should allow the research community to produce constantly improving, human curated atlases of the development of the ferret brain, but also of other species. The application will be based on the collaborative methodology that we have developed for our projects BrainSpell (http://brainspell.org) and Brain Catalogue (http://braincatalogue.org). As those projects, it will be based on open source technology, and its code released open source.
Implementation
Work plan
Our proposal is organised into 3 Work Packages and 4 milestones (Fig.
The data acquisition milestone and deliverables (part of WP 1) will be achieved by the end of the first year. Thanks to the optimised pipeline implemented by Neuroscience Associates, the sectioning and staining of the 16 brains should be done in less than 2 months, including shipping. Most of the time will be spent in scanning (Task T1.4) and data processing (Task T1.5). Scanning will be performed with a Zeiss AxioScan.Z1, able to process 50 double-slices per batch. We expect to process ~1,000 double-slices (each containing 4 to 16 individual brain slices), which will require 80 days of work and ~20 TB of data storage, plus backup.
The neuroinformatic work package (WP 2) will start at the beginning of the project, based on the MRI data that we have already acquired and continuing the development of our current tools. The work will be initially directed towards the harmonisation of the developments already achieved by the different partners, and will be later aimed at the construction and testing of a common platform. Our aim will be to start testing our visualisation tools with ferret data at the beginning of the second year, aiming at an early release of the visualisation tools 6 months later, and an early release of the collaboration tools at the beginning of the third year.
Finally, Work Package 3 will aim at coordinating the work of the partners, organising the resources necessary for collaboration (project website, common Github project, discussion list, meetings), the dissemination of our results (presentation at meetings, preparation of scientific papers), and the redaction of a report at the end of each year, summarising our results and perspectives.
Work packages
Table
Work Package 1. Histology production and scanning.
|
WP1 |
Histology production and scanning |
m1 |
m12 |
||||||
|
Contribution of project partners |
|||||||||
|
Partner number |
1 |
2 |
3 |
4 |
|||||
|
Total effort per partner (Person*months) |
20 |
8 |
|||||||
|
Aim of the WP
|
|||||||||
|
Tasks |
|||||||||
|
T1.1 |
Tissue preparation and shipping (m1: Responsible: 1; Involved: 2) Partner 1 (Pasteur) will prepare the brains for shipment to Neuroscience Associates. |
||||||||
|
T1.2 |
Sectioning (m2-m3: Responsible: 2; Involved: 1) Partner 2 (CEA) will work together with Neuroscience Associates to ensure the quality of the sectioning and block-face photographies. |
||||||||
|
T1.3 |
Staining (m2-m3: Responsible: 2; Involved: 1) Neuroscience Associates will stain the brain slices, condition them in double-slice boxes and ship them to Partner 1 (Pasteur), under the supervision of Partner 2 (CEA). |
||||||||
|
T1.4 |
Scanning (m3-m12: Responsible: 1; Involved: 2) Partner 1 (Pasteur) will organise a daily transfer of batches of slides to the scanner, organise the slides and launch an overnight scanning. Transfer the previously acquired the data for long-term storage. |
||||||||
|
T1.5 |
Data pre-processing and storage (m3-m12: Responsible: 1; Involved: 2, 4) At the beginning of the task, Partner 1 will organise in collaboration with Partners 2 (CEA) and 3 (McGill) the long-term storage of the data, and the preparation of scripts for the extraction of individual brains from the double-slices. Throughout the duration of the WP, Partner 1 will transfer the raw data to long-term storage, run and verify the extraction scripts. |
||||||||
|
Deliverable |
Month of delivery |
Title of deliverable |
|||||||
|
D1.1 |
3 |
Histology: sectioning and staining |
|||||||
|
D1.2 |
12 |
Histology data scanned, stored in CZI format, and backed up. |
|||||||
Table
Work Package 2. Data visualisation and collaboration platform.
|
WP2 |
Data vtform |
m1 |
m36 |
||||
|
Contribution of project partners |
|||||||
|
Partner number |
1 |
2 |
3 |
4 |
|||
|
Total effort per partner (Person*months) |
40 |
2 |
44 |
10 |
|||
|
Aims of the WP
|
|||||||
|
Tasks |
|||||||
|
T2.1 |
DeepZoom preprocessing, metadata and storage (m3-m14; Responsible: 1; Involved: 2, 4) Partner 1 will organise the data transfer and storage in collaboration with Partners 2 and 4. The data will be processed in regular batches at the High Performance Computing platform of the Institut Pasteur, and stored in parallel at HPB neuroinformatic platform. Partner 1 will compile meta-data for the deep-zoom data sets, containing information about quality assessments, resolution, etc. |
||||||
|
T2.2 |
Data visualisation tools (m1 – m18; Responsible: 2; Involved: 1, 4) Partner 1 in collaboration with Partners 3 and 4 will develop the web scripts for the organisation of the brain data, and the development of deep-zoom visualisation tools for multi-modal histology data and vectorial overlays (atlas, annotations). The code will be developed using github (http://github.com), that will be also used for discussion and tracking issues. Preliminary releases of the code will be pushed to github since the beginning of the task, and a first official release of the code (v1) will be pushed at the end of the task. Web hosting and data transfer to HBP will be organised by Partners 1 and 3. |
||||||
|
T2.3 |
Testing and Debugging (m18-m36; Responsible: 1; Involved: 3, 4) Our early release version will allow users (in addition to ourselves) to test the visualisation tools and inform us of bugs or required features through Github. We will deal with these changes until the end of the project, with the aim of providing a stable V1 version of the visualisation tools at the end of the project. |
||||||
|
T2.4 |
Web design (m15–m18; Responsible partner: 1; Involved partner: 3, 4) Partner 1 will organise the graphic design of the web site interface, and the user interactivity during the last 3 months of the project, to guarantee a high graphic quality, multi-platform (desktop/mobile) operability, and diffusion. The front-end and the back-end of the website will be made open-source and pushed to github. |
||||||
|
T2.5 |
Collaboration tools (m1-m24; Responsible: 1; Involved: 3, 4) Partners 1, 3 and 4 will survey and evaluate the open-source alternatives currently available, in particular, regarding the harmonisation of the applications already developed (Partner 1: BrainSpell, Brain Catalogue, Virtual microscope; Partner 3: Scalable Brain Atlas; Partner 4: BigBrain, BrainBrowser). The code will be developed using Github (http://github.com), that will be also used for discussion and tracking issues. Preliminary releases of the code will be pushed to github since the beginning of the task, and a first official release of the code (v1) will be pushed at the end of the task. |
||||||
|
T2.6 |
Testing and Debugging (m24-m36; Responsible: 1; Involved: 3,4) The early realise of the Collaboration tools should lead to bug reports and feature requirements from the users that will be addressed within this task. |
||||||
|
T2.7 |
Web design (m22-m24; Responsible partner: 1) Partner 1 will organise web development/desig during the last 2 months of the project, to guarantee a high graphic quality, multi-platform (desktop/mobile) operability, and diffusion. The front-end and the back-end of the website will be made open-source and pushed to github. |
||||||
|
Deliverable |
Month of delivery |
Title of deliverable |
|||||
|
D2.1 |
14 |
Data organised and stored in DZI format |
|||||
|
D2.2 |
18 |
Early release of the data visualisation tools |
|||||
|
D2.3 |
24 |
Early release of the collaboration tools |
|||||
|
D2.4 |
36 |
V1 of the collaboration code pushed to Github |
|||||
Table
Work Package 3. Coordination and dissemination.
|
WP3 |
Coordination and dissemination |
m1 |
m36 |
||||
|
Contribution of project partners |
|||||||
|
Partner number |
1 |
2 |
3 |
4 |
|||
|
Total effort per partner (Person*months) |
10 |
3 |
|||||
|
Aim of the WP The aim of this work package is to ensure the coordination of the different partners, facilitate the interaction, and the development of the project by setting up a clear organizational structure to monitor the progress of the project towards its planned objectives. In addition, in the framework of this WP, we will institute an effective dissemination plan to facilitate the transfer of knowledge among the beneficiaries and outside the consortium Partner 1 (Pasteur), as the Coordinating Institution, will handle the overall management of the FIIND project. To this end, Partner 1 will set up the resources necessary for collaboration: a project website, a common github repository, a common calendar, a discussion list, as well as the organisation of meetings (virtual/real-life). The coordinator will be responsible for collecting the necessary information from the beneficiaries to prepare the periodic and final scientific reports. It will also ensure the diffusion of our results, presentation at international meetings, outreach, preparation of scientific papers, by all involved partners. We envision the publication of a minimum of two papers, one describing the dataset (for example, for the journal Scientific Data), and another describing the neuroinformatic platform (for example, for the journal Nature Methods). Preprints of the papers produced during the project will be deposited at BioRxiv (http://biorxiv.org) to ensure a fast dissemination of the results, as well as to obtain an immediate feed-back from the community. We will present our projects at the annual conference of the Organisation for Human Brain Mapping. Finally, Partner 1 will coordinate the redaction at the end of each year a report summarising the results, difficulties encountered, and perspectives. A Consortium Agreement, covering such issues as the internal governance of the consortium, the management of Intellectual Property and access rights to results, liability and confidentiality arrangements between beneficiaries, will be drawn up by the Coordinating Institution and submitted for approval and signature by all participating institutions. The results arising from the project (Foreground) will be released under Non-attribution, Creative Commons license, taking into consideration the needs and expectations of the partners. |
|||||||
|
Tasks |
|||||||
|
T3.1 |
Implementation of the project management structure (m1 – m3; Responsible: 1; Involved: 2, 3, 4) Partner 1 will setup the resources necessary for collaboration, in collaboration with Partners 2- 4. The management of the FIIND project requires the implementation of an effective internal communication and management structure. |
||||||
|
T3.2 |
Dissemination and outreach (m1-m36; Responsible: 1; Involved: 2, 3, 4) Partner 1 will coordinate the diffusion of results and outreach. |
||||||
|
T3.3 |
Reports (m12, m24 and m36; Responsible: 1; Involved: 2, 3, 4) Partner 1 will will collect the necessary information to submit a scientific report on each 12- month period of the project. |
||||||
|
Deliverable |
Month of delivery |
Title of deliverable |
|||||
|
D3.1 |
12 |
First scientific report |
|||||
|
D3.2 |
24 |
Second scientific report |
|||||
|
D3.3 |
36 |
Final scientific report |
|||||
Table
Work package overview: Total effort per WP and partner (person-months)
|
Project-partner |
WP1 |
WP2 |
WP3 |
total |
|
1 Pasteur |
20 |
40 |
10 |
70 |
|
2 MIRCen |
8 |
2 |
10 |
|
|
3 Radboud |
44 |
3 |
47 |
|
|
4 McGill |
10 |
10 |
||
|
total |
28 |
96 |
13 |
137 |
Management and risk assessment
Table
List of milestones
|
No of Milestone |
Delivery month |
WP involved |
Title |
|
M1 |
12 |
WP1 |
Data acquired, scanned and stored |
|
M2 |
18 |
WP2 |
Data made public, visualisation tools early release |
|
M3 |
24 |
WP2 |
Collaborative tools early release |
|
M4 |
36 |
WP2 |
Release of V1 visualisation and collaborative tools |
The project is coordinated by Partner 1 (Pasteur), who is also responsible for Work Packages 1 (data acquisition) and 2 (neuroinformatics development).
Work Package 1 includes the histological processing of 16 brains. Our project depends critically on histology data acquisition, which is the most important bottleneck for Work Package 1. Work Package 2, neuroinformatic development, also requires the histology data from WP1, however, during all the 1st year it can progress independently. To ensure that the quality of data acquisition and timing will be respected, we will work in collaboration with Neuroscience Associates. Neuroscience Associates focus on high-throughput, reliable histology processing. Together with Partner 2, they have developed and implemented an optimised pipeline that allows them to produce high-quality, homogeneous, reproducible results. They have not worked with ferrets before, but they have extensive experience with mice and rats, and have also processed successfully large brains such as those of dogs, pigs and bears, among many others.
Our project should produce ~12,000 individual brain slices, that need to be digitised before they could be processed and used by WP 2. Here, again, relying on the MultiBrain technology of Neuroscience Associated will simplify the task. Because all 16 brains will be processed at the same time, and fixated on a regular grid of 16 brains per double slide, the scanning time will be reduced and the extraction of the individual brains from the double slides simplified. The histology platform where the slides will be scanned is in the computer network of the Institut Pasteur, and the data will be directly transferred from where it is acquired to the informatic platform, where it will be stored and processed. Partner 1 is physically located within the campus of the Institut Pasteur, and will be able to coordinate the correct progress of the tasks. This should allow us to complete Milestone 1, the acquisition, scanning and storage of the data by the end of the 1st year of the project.
Work Package 2 comprises all neuroinformatic development, for visualisation and collaborative analysis of the data. During the 1st year of the project, WP 2 does can progress without the final data, towards the refinement and development of visualisation and collaboration tools. The responsible for WP 2 is Partner 1 (Pasteur) in collaboration with Partners 3 (Radboud) and 4 (McGill). In our prototype stage we have successfully used the cell-scale resolution Nissl stained data from the Allen Institute, and Partner 4 has also access to cell-scale resolution data from their BigBrain project. The ability to collaborate smoothly among the 3 partners involved is critical. We will rely on Github for collaboration, which is a widely used portal for collaborative development of software, including revision control, source code management, wikis, task management, bug tracking and feature requests. Finally, we will work in synergy with members of HBP to ensure the integration and interoperability with NIP tools.
Consortium
Partner 1. Institut Pasteur, Department of Neuroscience, Project coordinator: Roberto Toro
Expertise of the organisation:
The Department of Neuroscience at the Institut Pasteur of Paris aims to become a world leader in the elucidation of the genetic (and epigenetic), molecular and cellular mechanisms underlying the neural basis of behaviour and social interactions, and brain alteration in neurological and psychiatric disorders. Research programs within the department are fundamental in nature, but are expected to provide a better understanding of human brain disease and identify new therapeutic approaches. The Institut Pasteur provides to the researchers in its campus access to several scientific platforms. For the present project, we will have access to the bioinformatics platform (high-performance computing, data storage), and to the histology platform (high-throughput, high-resolution scanning).
Principal investigator:
Roberto Toro is group leader at the Department of Neuroscience of the Institut Pasteur. After a degree in engineering, he obtained a Master and a PhD in Neuroscience at the University of Paris 6. Roberto Toro is interested on the development and evolution of the brain, which he studies through mathematical modelling, magnetic resonance imaging and genomics. The aim of his research is to develop a theoretical framework to understand and analyse neuranatomical data, its normal variability, and its relationship with neurodevelopmental disorders. In his theoretical work, he has developed computational models to describe the global organisation of the sulcal anatomy of the human brain, and to simulate the development of neocortical folding. In computational neuroanatomy, he has developed methods to measure cortical folding, as well as the non-linear patterns of neuroanatomical variability. In genomics, he is interested in the way in which genetic and environmental factors determine neuroanatomical diversity. He has investigated the genomic architecture of human neuroanatomical diversity, and participated in the analysis of the genetic bases of human neuroanatomy of the ENIGMA consortium, being part of the analysis team. The work of Roberto Toro also aims at better understanding the interaction between normal diversity and pathology in different neurological disorders, and he is especially interested in the genetics and neuroanatomical changes associated with autism spectrum disorders.
Publications:
- Toro R, Poline JB, Huguet G, Loth E, Frouin V, Banaschewski T, et al (2014) Genomic architecture of human neuroanatomical diversity. Mol Psychiatry, doi: 10.1038/mp.2014.99
- Delorme R, Ey E, Toro R, Leboyer M, Gillberg C, Bourgeron T (2013) Progress toward treatments for synaptic defects in autism. Nature Medicine 19 (6), 685-694
- JL Stein, SE Medland, A Arias Vasquez, DP Hibar, RE Senstad, AM Winkler, R Toro, et al (2012) Common genetic polymorphisms are associated with human hippocampal and intracranial volumes. Nature Genetics.
- Toro R (2012) On the possible shapes of the brain. Evol Biol, doi: 10.1007/s11692-012-9201- 8
- Kelly C, Toro R, Di Martino A, Cox CL, Castellanos FX, Milham MP (2012) Convergent Functional Architecture of the Insula Emerges Across Imaging Modalities. NeuroImage, doi: 10.1016/j.neuroimage.2012.03.021
Role in project:
- Project coordination
- Scanning of histological data
- Computational neuroanatomy and neuroinformatics: MRI and histology data pre-processing,
- data analysis, development of web tools for data visualisation, creation of collaborative atlas, coordination of web development.
Partner 2. Commissariat à l'Énergie Atomique (French Atomic Commission), Molecular Imaging Research Centre (MIRCen), France, Thierry Delzescaux
Expertise of the organisation related to the project objectives.
The MIRCen (Molecular Imaging Research Center) is a preclinical research centre which brings together several technological platforms dedicated to the development of relevant animal models of human diseases and to the evaluation of novel therapies, primarily in the field of neurodegenerative diseases. These facilities, spread over 8500 m2, possess a high concentration of complementary expertise and methodologies. In vivo (PET, MRI) and post mortem (histology, microscopy) imaging techniques play a key role to support these researches. In particular, methodological developments concerning 3D post mortem reconstruction and analysis of rodent and primate brains have been performed during the last decade as well as in vivo/post mortem multimodal 3D co-registration.
Principal investigator Thierry Delzescaux: PhD image processing, CEA researcher
- Image processing, specialisation in preclinical research (experimental models),
- Image processing in neurodegenerative diseases (Alzheimer's, Parkinson's, Huntington's),
- Multimodal co-registration of in vivo (PET, MRI, CT) and post mortem (histology, autoradiography, immunohistochemistry) brain images, 3D reconstruction and image analysis of post mortem / ex vivo data, anatomo-functional studies in rodents and primates,
- Coordinator of BrainRAT project (Brain Reconstruction and Analysis Toolbox) developed by the image processing team of the MIRCen and integrated into BrainVISA software (http://brainvisa.info).
Publications:
- Boussicault L., Hérard A.-S., Calingasan N., Petit F., Malgorn C., Merienne N., Jan C., Gaillard M.-C., Lerchundi R., Barros L.-F., Escartin C., Delzescaux T., Mariani J., Hantraye P., Beal M.-F., Brouillet E., Véga C., Bonvento G. (2014) Impaired brain energy metabolism in the BACHD mouse model of Huntington's disease: critical role of astrocyte-neuron interactions. J Cereb Blood Flow Metab, 34(9), 1500-1510.
- Damiano M., Diguet E., Malgorn C., D'Aurelio M., Galvan L., Petit F., Benhaim L., Guillermier M., Houitte D., Dufour N., Hantraye P., Canals J.-M., Alberch J., Delzescaux T., Déglon N., Beal M.-F., Brouillet E. (2013) A role of mitochondrial complex II defects in genetic models of Huntington's disease expressing N-terminal fragments of mutant huntingtin. Hum Mol Genet, 22(19), 3869-3882.
- Lavisse S., Guillermier M., Hérard A.-S., Petit F., Delahaye M., Camp N. V., Ben Haim L., Lebon V., RemyP., Dollé F., Delzescaux T., Bonvento G., Hantraye P., Escartin C. (2012) Reactive astrocytes overexpress TSPO and are detected by TSPO positron emission tomography imaging. J Neurosci, 32(32), 10809-10818.
- Lebenberg J., Hérard A-S., Dubois A., Dhenain M., Hantraye P., Delzescaux T. (2011) A combination of atlas-based and voxel-wise approaches to analyze metabolic changes in autoradiographic data from Alzheimer’s mice. NeuroImage 57: 1447–1457.
- Dubois A, Dauguet J, and Delzescaux T (2011) Ex vivo and in vitro cross calibration methods. In Fabian Kiessling and Bernd J. Pichler, editors, Small Animal Imaging, pages 317–346. Springer Berlin Heidelberg (book chapter).
Role in project:
- Coordination of histology production in collaboration with NSA company (protocol definition, sampling, staining, photographic modality).
- Image processing expertise and contribution: colour image segmentation, 3D post mortem reconstruction, in vivo/post mortem multimodal co-registration.
Partner 3. Radboud University Nijmegen, Neuroinformatics, Paul Tiesinga, Rembrandt Bakker.
Expertise:
The Neuroinformatics department of Donders Centre for Neuroscience currently has four major research themes. First, to understand processing of sensory information in the brain, and the modulation thereof by cognitive factors, at the level of networks of spiking neurons. This is accomplished by building biophysical models ranging in size from single neurons up to many hundred thousands of neurons that incorporate the latest physiological and anatomical data and by performing experiments. Second, to develop methods to analyse multivariate data recorded from the brain in order to extract and validate signs of communication between cortical networks.
Third, to develop and apply machine-learning methods in order to build predictive models for brain network structure, for instance, by utilizing data fusion techniques to combine data from different modalities and matrix completion methods to impute missing data. Fourth, to develop neuroinformatics infrastructures to organize and analyse neuroscience data of a variety of sources.
Principal investigator:
Paul Tiesinga is professor of Neuroinformatics and chair of the department of Neuroinformatics. He received his masters in Theoretical Physics (1992) as well as his PhD in physics (1996) from Utrecht University. He was a postdoc in the physics department at Northeastern University (1997-1998, Boston, US) and a Sloan postdoctoral fellow at the Salk Institute (1998-2002, La Jolla, US). In 2002, he became an assistant professor in the Physics & Astronomy department at the University of North Carolina in Chapel Hill and was in 2008 promoted to associate professor. In 2009, he moved to the Radboud University Nijmegen to establish the Neuroinformatics department. He served as director of the Donders Centre for Neuroscience from 2010 to 2014 and is member of the Governing Board of the International Neuroinformatics Facility (INCF) since 2012. He has supervised 5 postdocs, 2 PhD students, and is currently supervising 2 postdocs, 3 PhD students and 5 master students and in addition co- supervising 7 PhDs.
Co-Investigator:
Rembrandt Bakker is an experienced postdoc who joined the Neuroinformatics department in 2009, and built up the Scalable Brain Atlas (https://scalablebrainatlas.incf.org) from scratch. He has been closely involved in the INCF digital atlasing infrastructure project. He has also demonstrated his capacities as an eScience engineer in the development and curation of two other ICT projects:
1. The CoCoMac database of macaque structural connectivity, a long-established resource the uses a well defined ontology for the curation and processing of manually collated tract-tracing study publications (http://cocomac.g-node.org).
2. The Biomarker Boosting eScience infrastructure, a platform for the storage and processing of privacy-sensitive medical imaging data. (http://ww.esciencecenter.nl/project/biomarker-boosting).
Publications:
- Bakker, R., Tiesinga, P. & Koetter, R. (2015) The Scalable Brain Atlas: instant web-based access to public brain atlases and related content. Neuroinformatics.
- Bezgin, G., Vakorin, V.A., van Opstal, A.J., McIntosh, A.R. & Bakker, R. (2012) Hundreds of brain maps in one atlas: registering coordinate-independent primate neuro-anatomical data to a standard brain. NeuroImage, 62, 67-76.
- Sergejeva, M., Papp, E.A., Bakker, R., Gaudnek, M.A., Okamura-Oho, Y., Boline, J., Bjaalie, J.G. & Hess, A. (2015) Anatomical landmarks for registration of experimental image data to volumetric rodent brain atlasing templates. Journal of neuroscience methods, 240, 161-169.
- Tiesinga, P. & Sejnowski, T.J. (2009) Cortical enlightenment: are attentional gamma oscillations driven by ING or PING? Neuron, 63, 727-732.
- Womelsdorf, T., Valiante, T.A., Sahin, N.T., Miller, K.J. & Tiesinga, P. (2014) Dynamic circuit motifs underlying rhythmic gain control, gating and integration. Nature neuroscience, 17, 1031-1039.
Role in project:
- Registration of high resolution slice data to medium resolution block-face images
- Development of the virtual microscopy web service
- Development of the platform for collaborative atlases building
- Integration of visualisation/collaboration services with the HBP Unified Portal and Scalable Brain Atlas
Partner 4. McGill University, McGill Centre for Integrative Neuroscience, Alan Evans.
Expertise:
The McGill Centre for Integrative Neuroscience, directed by Professor Alan Evans, has developed many important technologies such as LORIS, a web-accessible database solution for Neuroimaging; CBRAIN, a collaborative, web-enabled grid platform that allows researcher to perform computationally expensive analyses on distributed data; BrainBrowser, a WebGL-based 3D viewer for displaying 3D surfaces of brain in real-time and superimposing various data to those surfaces; the MINC tools and the MINC and MINC2 file formats (based on NetCDF data format), and CIVET, an automated human brain-imaging pipeline for preprocessing, quality control and analysis of large MR data sets.
Principal Investigator:
Professor Alan Evans was originally trained in physics at Liverpool University in the U.K. He completed his PhD in biophysics, studying the 3D folding patterns of protein structure and the binding of co-factors and substrates to enzymes. He then spent 5-year at Atomic Energy of Canada Ltd. in Ottawa, working on the physics and biochemical analysis of positron emission tomography (PET) data. In 1984, he moved to the Montreal Neurological Institute (MNI) at McGill University in Montreal to continue his PET research. His research interests include multi-modal brain imaging with PET and MRI, image processing and large-scale brain database analysis.He has published over 300 peer-reviewed papers and holds numerous NIH and CIHR grants. During his 25 years at the MNI, he has held numerous leadership roles, most notably as director of the McConnell Brain Imaging Centre (BIC) during the 1990’s. Dr. Evans is a founding member of the International Consortium for Brain Mapping (ICBM). He was one of the founders of the Organization for Human Brain Mapping (OHBM), serving in numerous positions on the OHBM Council since 1995. He chaired the 4th International Conference on Human Brain Mapping in 1998. He is a regular participant in numerous NIH workshops, panels and initiatives related to brain imaging research. He is on the scientific advisory board of numerous research programs in this field. In 2003 he received a prestigious Senior Scientist Award from the Canadian Institutes of Health Research.
Publications:
- JJ Wolff et al (2014) Differences in white matter fiber tract development present from 6 to 24 months in infants with autism. American Journal of Psychiatry 169 (6), 589-600
- K Amunts et al (2013) BigBrain: an ultrahigh-resolution 3D human brain model. Science 340 (6139), 1472-1475
- J Chen et al (2012) Maternal Deprivation in Rats is Associated with Corticotrophin–Releasing Hormone (CRH) Promoter Hypomethylation and Enhances CRH Transcriptional Responses to Stress in Adulthood. J Neuroendocrinology 24 (7), 1055-1064
- P Shaw et al (2012) Development of cortical surface area and gyrification in attention- deficit/hyperactivity disorder. Biological psychiatry 72 (3), 191-197
- AC Evans et al (2012) Brain templates and atlases. Neuroimage 62 (2), 911-922
Role in project:
- Development of data visualisation tools
- Development of on-line collaboration tools
Please explain how the partner is able to secure its own funding
Alan Evans currently holds numerous NIH and CIHR grants, specifically from Brain Canada (2014- 2019), CIHR Operating Grant (2013-2018), Cbrain grant (2013-2016), Catalyst grant (2014-2016), Autism Centers of Excellence (ACE) Networks Grant (2012-2017). From these grants he supports a team of ten developers for the LORIS and CBrain platforms. These developers will share their code and knowledge and contribute to the visualisation and collaboration tools.
Our consortium combines an international team of researchers with complementary expertises. All four partners share a background in computational neuroanatomy. Within this common context, Partners 1 and 3 have worked on different aspects of mathematical modelling: neurodynamic models of brain function in the case of Partner 3 and mechanical models of neocortical development in the case of Partner 1. Partners 2, 3 and 4 have worked in the integration of histological and MRI data: Partner 2 has a long experience in the registration of histological/MRI data in rodents and primates, Partner 3 has implemented a multi-atlas registration pipeline to automatically segment brain regions, and Partner 4 is one of the leaders of the BigBrain project, combining histology/MRI data for the human brain. Partners 1, 3 and 4 share an interest in online data visualisation, and have already developed complementary technologies for the visualisation of MRI data, surface reconstructions and atlases. Finally, all partners are engaged and promote data sharing and open, collaborative science.
Our project will provide a direct contribution to international and European research, by making freely available a valuable dataset and the neuroinformatics tools required to access, visualise and exploit it.
Consortium agreement principles (partner’s rights and duties, IPR management)
The Consortium Agreement will describe the administrative and the legal management of the project and will define provisioning measure on: management and organisational provisions (committees, conflict resolution procedures), legal provisions and management of IPR.
Important components of intellectual property (IP) (confidentiality, joint ownership, transfer of project results, access rights) will be specified in the consortium agreement. The information and IP rights each participant holds before entering the project (e.g. procedures and technologies) that participants wish to exclude from access will be identified at an early stage and attached as annex to the consortium agreement. Details of how intellectual property will be protected will be part of the agreement elaborated between the participants after approval of the proposal. Intellectual property issues will be coordinated by the Partner 1.
The consortium and the coordinator will monitor the visibility of research output and publication of results in peer-reviewed journals. The coordinator will inform and emphasise the importance of Open Access throughout the project lifetime.
Description of significant facilities and large equipment available to the consortium to perform the project
- High-performance computing facility, ~1300 computing nodes (to be increased to ~3000 in the next months) (Partner 1).
- High-throughput histology slice scanner, Zeiss AxioScan.Z1. Accepts up to 25 double-slides, for scanning at a magnification factor of x20 (Partner 1).
- Data storage, backup and network. The histology scanner and the HPC facility are connected to the same high-speed network, which facilitates data integration and processing (Partner 1).
- The Neuroinformatics department one 8 core workstation available as a development machine to jump-start the project. It also has a share in the high performance computing cluster at the faculty of science at Radboud University Nijmegen, which comprises 1324 cores and 7914 GB RAM (Partner 3).
Link with ongoing projects
- Partner 1, Brain Catalogue. Project e-Museum, 150k€ from the Muséum National d'Histoire Naturelle de Paris, France, to establish an online, collaborative database of vertebrate brain MRI from ~2,000 specimens.
- Partner 1, Brain Catalogue. Project e-Museum, 50k€, from the Muséum National d'Histoire Naturelle de Paris, France, for web development and web design associated with the Brain Catalogue project.
- Partner 3 is leader of the predictive neuroinformatics Workpackage within the Human Brain Project and develops machine-learning based approaches to predict missing connectivity data. Partner 3 is thereby strongly embedded in the neuroinformatics subproject and forms the bridge between structural analysis work package of Chema Peña, the atlasing workpackage of Jan Bjaalie, and the neuroinformatics data integrator Martin Telefont.
- Partner 3 is also the coordinator of the Biomarker Boosting grant, funded through a collaboration between the Netherlands eScience Center and the Netherlands foundation for scientific research (NWO), and which has as aim to combine neuroimaging data sets acquired during different studies at four different university medical centers. Key components of the project are: data sharing, image processing of neuroimaging data and the statistical analysis thereof.
Financial Plan
- Partner 1. We have a quotation from Neuroscience Associates (NSA) for the high-throughput 3D histology processing, slicing and staining, of 67k€. Working with NSA is important to obtain a homogeneous and reliable dataset, within the short duration of our project. Three people will work in this project: Roberto Toro, group leader, 20 person months (117,492€, funded by the Institut Pasteur); Ophélie Foubet, PhD student, 24 person months (95,450€, funded by the Centre de Recherche Interdisciplinaire de Paris); and we require funding for 1 year of experienced post-doctoral contract (57,646€) to assist with the last phases of neuroinformatic development. The profile for this postdoctoral contract will be computational neuroanatomy, neuroinformatics, experience with online development and data visualisation. We also request funding for consumables (Network Accessible Storage for fast data storage and data backup: 20k€), publication fees (5k€), travel fees (2 meeting per year for 2 people, 8k€). Adding administrative fees, the total amount requested by Partner one is 163,952€. The amount requested for subcontracting (67k€ for Neuroscience Associates), represents less than 50% of the total amount requested to the Agence Nationale pour la Recherche (ANR, France), in agreement with their conditions.
- Partner 3. Two researchers will contribute to this project in kind: Paul Tiesinga, Head of the Neuroinformatics department (5% effort (1.8PM), 21,000 €, funded by Radboud university); Rembrandt Bakker (10% effort (3.6PM) €24,000, funded through the HBP project); and we require funding for two additional people, 3 year of experienced post-doctoral researcher (36PM, €208.670 according to the NWO pay scale, starting with 2 years experience) to develop the Virtual microscopy web portal and the Platform for building collaborative atlases, during year 2 we will need a programmer at the Masters’ level (6PM, €23840, NWO pay scale for PhD student with 2 years experience) to deploy the first version of the portal/platform. The profile for the postdoctoral researcher will be computational neuroanatomy, neuroinformatics, experience with online development and data visualisation. The programmer will need experience with standard programming languages. We also request funding for consumables (software packages, data storage and nodes for our HPC cluster 7.790k€), publication fees (2k€), travel costs (2 meeting per year for 2 people, 7.5k€). The total amount requested by Partner one is 250K€.
Impact
Expected Impacts
The FIIND project will produce two main concrete outcomes: an open-access, integrated and interactive atlas of Ferret brain development, and a set of species-agnostic web services for the collaborative, online, construction of multi-species atlases.
1. Atlas of Ferret brain development. The raw dataset will be made available by month 12 the project. It will contain up to 12,000 high-resolution slices of 16 ferret brains at age P0, P4, P8 and P16. The dataset will contain stainings for cytoarchitectonic landmarks and neurogenesis. The data will be made available through HBP's Unified Portal. This resource will provide the researcher community with a high-quality dataset to investigate the relationship between multiple scales of brain organisation, across developmental stages.
2. Neuroinformatics tools. We plan a first early release of the integrated, interactive data visualisation platform by month 18 of the project; and an early release of the collaborative atlas framework by month 24 of the project. During the last year of the project we will focus on the consolidation of the project, issue tracking, and bug correction, so that Version 1 of the neuroinformatics tools will be made available at the end of the project, month 36. The development will be done online, and all tools will be made available, open-source, through Github.
The HBP focuses primarily on constructing models for the mouse brain and for the human brain. The results of our project will provide HBP researchers with all the necessary data and neuroinformatics tool to investigate and model the brain of an intermediate species, the ferret. The dataset and tools will be released in open access, making sure that our results are re-usable (that they are not bound to answer a single question or hypothesis), and interoperable (that data is provided in open, standard formats, so that it can be accessed with other tools). In particular, we will make sure that our pipeline and data formats are compatible and integrated with HBP's Neuroinformatics Platform.
Dissemination and exploitation of results
Effective dissemination and use of results is greatly important for a pan-European initiative such as FIIND which aims to provide a detailed description of the development of a complex brain. Given the expected impacts which could lead to a better understanding of brain development, the nature of human neuroimaging data, the creation of models of brain development and the production of the first integrated atlas of ferret brain development, FIIND will emphasize dissemination of results to the scientific community and to the public to ensure exploitation of its results. In our external dissemination and exploitation of results activities we will distinguish two main target audiences, each requiring a different approach:
Towards the research community:
- Scientific publications and international conferences: The coordinator will monitor the visibility of research output and publication of results in peer-reviewed journals, and at international conferences.
- FIIND web platform: The web platform will allow researchers to access and visualize interactively the MRI, histology and 3D reconstruction data.
- Open Access: The coordinator will inform and emphasize, throughout the project lifetime, the importance of Open Access to ensure all partners are aware of the practice of providing on- line access to scientific information that is open and free of charge to the end-user.
Towards the General public:
- The transparency and accessibility of the project to the general public is of great importance to ensure EU citizens are informed of FIIND aims, developments and outcomes. The public website will be an important component of this strategy.
- Publication in the lay press will be encouraged to attract maximum attention on the project.
- “Brain Awareness Week”: Each year is celebrated in Europe the “Brain Awareness Week” to increase public awareness of the progress and benefits of brain research. Our consortium will actively participate in the preparation of this event to inform on the outcomes of the project and present our collaborative atlas of ferret brain development.
- Citizen science and crowdsourcing are emerging as important new strategies to involve directly the general public into research. As we will encourage the collaboration among scientists, we will also encourage the discussion and participation of the general public in our research, through the participation and organisation of crowdsourcing contests and hackatons.
Ethical Issues
Ferrets were kept at the Animal Facilities of the Universidad Miguel Hernández, where animals were treated according with Spanish and EU regulations and experimental protocols were approved by the Universidad Miguel Hernández IACUC.
Funding program
FLAG-ERA Joint Transnational Call (JTC) 2015
Hosting institution
Institut Pasteur, Paris, France; Radboud University Nijmegen, The Netherlands
References
-
BigBrain: An Ultrahigh-Resolution 3D Human Brain Model.Science340:1472‑1475. https://doi.org/10.1126/science.1235381
-
Characterization of Brain Development in the Ferret via MRI.Pediatr Res66:80‑84. https://doi.org/10.1203/PDR.0b013e3181a291d9
-
Development of the human cerebral cortex: Boulder Committee revisited.Nat Rev Neurosci9:110‑22. https://doi.org/10.1038/nrn2252
-
Mapping the early cortical folding process in the preterm newborn brain.Cereb Cortex N Y N18:1444‑54. https://doi.org/10.1093/cercor/bhm180
-
Neurogenic radial glia in the outer subventricular zone of human neocortex.Nature464:554‑561. https://doi.org/10.1038/nature08845
-
The Allen Brain Atlas.Springer Handb Bio-NeuroinformaticsSpringer Berlin,Heidelberg.
-
Exuberance in the development of cortical networks.Nat Rev Neurosci6:955‑965. https://doi.org/10.1038/nrn1790
-
Evolution of columns, modules, and domains in the neocortex of primates.Proc. Natl. Acad. Sci109:10655‑10660. https://doi.org/10.1073/pnas.1201892109
-
Regional Patterns of Cerebral Cortical Differentiation Determined by Diffusion Tensor MRI.Cereb Cortex19:2916‑2929. https://doi.org/10.1093/cercor/bhp061
-
Area patterning of the mammalian cortex.Neuron56:252‑69. https://doi.org/10.1016/j.neuron.2007.10.010
-
The Rat Brain in Stereotaxic Coordinates: Compact, Sixth Edition.6.Academic Press
-
Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates, Fourth.Academic Press,Amsterdam.
-
Evolution of the neocortex: a perspective from developmental biology.Nat Rev Neurosci10:724‑735. https://doi.org/10.1038/nrn2719
-
A Role for Intermediate Radial Glia in the Tangential Expansion of the.Mammalian Cerebral Cortex. Cereb Cortex21:1674‑1694. https://doi.org/10.1093/cercor/bhq238
-
Development of cerebral sulci and gyri in ferrets (Mustela putorius).Congenit Anom52:168‑175. https://doi.org/10.1111/j.1741-4520.2012.00372.x
-
Gyrus formation in the cerebral cortex in the ferret. I. Description of the external changes.J Anat146:141‑152.
-
Gyrus formation in the cerebral cortex of the ferret. II. Description of the internal histological changes.J Anat147:27‑43.
-
Co-Planar Stereotaxic Atlas of the Human Brain: 3-D Proportional System: An Approach to Cerebral Imagingedition. Stuttgart.1.Thieme,New York.
-
Why Does Cerebral Cortex Fissure and Fold?Cerebral Cortex8:3‑136. https://doi.org/10.1007/978-1-4615-3824-0_1
-
Primate Brain Maps: Structure of the Macaque Brain: A Laboratory Guide with Original Brain Sections, Printed Atlas and Electronic Templates for Data and Schematics (including CD-ROM).Elsevier Science,New York.
-
The Analysis of Cortical Connectivity.1.Bioscience,New York; Austin: Landes.
-
A universal scaling law between gray matter and white matter of cerebral cortex.Proc. Natl. Acad. Sci97:5621‑5626. https://doi.org/10.1073/pnas.090504197
-
Centenary of Brodmann’s map — conception and fate.Nat Rev Neurosci11:139‑145. https://doi.org/10.1038/nrn2776