|
Research Ideas and Outcomes : Guidelines
|
|
Corresponding author: Lyubomir Penev (penev@pensoft.net)
Received: 26 Feb 2017 | Published: 28 Feb 2017
© 2018 Lyubomir Penev, Daniel Mietchen, Vishwas Chavan, Gregor Hagedorn, Vincent Smith, David Shotton, Éamonn Ó Tuama, Viktor Senderov, Teodor Georgiev, Pavel Stoev, Quentin Groom, David Remsen, Scott Edmunds
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Penev L, Mietchen D, Chavan V, Hagedorn G, Smith V, Shotton D, Ó Tuama É, Senderov V, Georgiev T, Stoev P, Groom Q, Remsen D, Edmunds S (2017) Strategies and guidelines for scholarly publishing of biodiversity data. Research Ideas and Outcomes 3: e12431. https://doi.org/10.3897/rio.3.e12431
|
|
Abstract
The present paper describes policies and guidelines for scholarly publishing of biodiversity and biodiversity-related data, elaborated and updated during the Framework Program 7 EU BON project, on the basis of an earlier version published on Pensoft's website in 2011. The document discusses some general concepts, including a definition of datasets, incentives to publish data and licenses for data publishing. Further, it defines and compares several routes for data publishing, namely as (1) supplementary files to research articles, which may be made available directly by the publisher, or (2) published in a specialized open data repository with a link to it from the research article, or (3) as a data paper, i.e., a specific, stand-alone publication describing a particular dataset or a collection of datasets, or (4) integrated narrative and data publishing through online import/download of data into/from manuscripts, as provided by the Biodiversity Data Journal.
The paper also contains detailed instructions on how to prepare and peer review data intended for publication, listed under the Guidelines for Authors and Reviewers, respectively. Special attention is given to existing standards, protocols and tools to facilitate data publishing, such as the Integrated Publishing Toolkit of the Global Biodiversity Information Facility (GBIF IPT) and the DarwinCore Archive (DwC-A).
A separate section describes most leading data hosting/indexing infrastructures and repositories for biodiversity and ecological data.
Keywords
biodiversity data publishing, data publishing licenses, Darwin Core, Darwin Core Archive, data re-use, data repository
Data Publishing in a Nutshell
Introduction
Data publishing in this digital age is the act of making data available on the Internet, so that they can be downloaded, analysed, re-used and cited by people and organisations other than the creators of the data (
- Data hosting, long-term preservation and archiving
- Documentation and metadata
- Citation and credit to the data authors
- Licenses for publishing and re-use
- Data interoperability standards
- Format of published data
- Software used for creation and retrieval
- Dissemination of published data
The present guidelines are based on an earlier version published in PDF on Pensoft's website in 2011 (
The FORCE11 group dedicated to facilitating change in knowledge creation and sharing, recognising that data should be valued as publisheable and citable products of research, has developed a set of principles for publishing and citing such data. The FAIR Data Publishing Group formulated the following four FAIR principles of fata publishing (
- Data should be Findable
- Data should be Accessible
- Data should be Interoperable
- Data should be Re-usable.
A key outcome of FORCE11 is the Joint Declaration of Data Citation Principles (see also
- Importance: Data should be considered legitimate, citable products of research. Data citations should be accorded the same importance in the scholarly record as citations of other research objects, such as publications.
- Credit and Attribution: Data citations should facilitate giving scholarly credit and normative and legal attribution to all contributors to the data, recognizing that a single style or mechanism of attribution may not be applicable to all data.
- Evidence: In scholarly literature, whenever and wherever a claim relies upon data, the corresponding data should be cited.
- Unique Identification: A data citation should include a persistent method for identification that is machine actionable, globally unique, and widely used by a community.
- Access: Data citations should facilitate access to the data themselves and to such associated metadata, documentation, code, and other materials, as are necessary for both humans and machines to make informed use of the referenced data.
- Persistence: Unique identifiers — and metadata describing the data and its disposition — should persist, even beyond the lifespan of the data they describe.
- Specificity and Verifiability: Data citations should facilitate identification of, access to, and verification of the specific data or datum that support a claim. Citations or citation metadata should include information about provenance and permanence sufficient to facilitate verfiying that the specific timeslice, version and/or granular portion of data retrieved subsequently is the same as was originally cited.
- Interoperability and Flexibility: Data citation methods should be sufficiently flexible to accommodate the variant practices among communities, but should not differ so much that they compromise interoperability of data citation practices across communities.
The Research Data Alliance (RDA) promotes the open sharing of data by building upon the underlying social and technical infrastructure. Established in 2013 by the European Union, the National Science Foundation and the National Institute of Standards and Technology (USA) as well as the Department of Innovation (Australia), it has grown to include some 4,200 members from 110 countries who collaborate through Work and Interest Groups "to develop and adopt infrastructure that promotes data-sharing and data-driven research, and accelerate the growth of a cohesive data community that integrates contributors across domain, research, national, geographical and generational boundaries" (
- Data Description Registry Interoperability Model
- Persistent Identifier Type Registry
- Workflows for Research Data Publishing: Models and Key Components
- Bibliometric Indicators for Data Publishing
- Dynamic Data Citation Methodology
- Repository Audit and Certification Catalogues
One RDA output, the Scholix Inititive, under the RDA/WDS (ICSU World Data System) Publishing Data Services Work Group is of particular relevance, as it seeks to develop an interoperability framework for exchanging information about the links between scholarly literature and data, i.e., what data underpins literature and what literature references data.
Within RDA, a Biodiversity Data Integration Interest Group has been established, which aims to "increase the effectiveness of biodiversity e-Infrastructures by promoting the adoption of common tools and services establishing data interoperability within the biodiversity domain, enabling the convergence on shared terminology and routines for assembling and integrating biodiversity data."
With regard to biodiversity, some recently published papers emphasise the importance of publishing of biodiversity data (
The EU BON project funded by the European Union's Framework Program Seven (FP7) (Building the European Biodiversity Observation Network, grant agreement ENV30845) was launched to contribute towards the achievement of these challenging tasks within a much wider global initiative, the Group on Earth Observations Biodiversity Observation Network (GEO BON), which itself is a part of the Group of Earth Observation System of Systems (GEOSS). A key feature of EU BON is the delivery of near-real-time data, both from on-ground observation and remote sensing, to the various stakeholders to enable greater interoperability of different data layers and systems, and provide access to improved analytical tools and services; furthermore, EU BON is supporting biodiversity science-policy interfaces, facilitate political decisions for sound environmental management (
The present paper outlines the strategies and guidelines needed to support the scholarly publishing and dissemination of biodiversity data, that is publishing through the academic journal networks.
What Is a Dataset
A dataset is understood here as a digital collection of logically connected facts (observations, descriptions or measurements), typically structured in tabular form as a set of records, with each record comprising a set of fields, and recorded in one or more computer data files that together comprise a data package. Certain types of research datasets, e.g., a video recording of animal behaviour, will not be in tabular form, although analyses of such recordings may be. Within the domain of biodiversity, a dataset can be any discrete collection of data underlying a paper – e.g., a list of all species occurrences published in the paper, data tables from which a graph or map is produced, digital images or videos that are the basis for conclusions, an appendix with morphological measurements, or ecological observations.
More generally, with the development of XML-based publishing technologies, the research and publishing communities are coming to a much wider definition of data, proposed in the BioMed Central (BMC) position statement on open data: "the raw, non-copyrightable facts provided in an article or its associated additional files, which are potentially available for harvesting and re-use" (
As these examples illustrate, while the term "dataset" is convenient and widely used, its definition is vague. Data repositories such as Dryad, wishing for precision, do not use the term "dataset". Instead, they describe data packages to which metadata and unique identifiers are assigned. Each data package comprises one or more related data files, these being data-containing digital files in defined formats, to which unique identifiers and metadata are also assigned. Nevertheless, the term "dataset" is used below, except where a more specific distinction is required.
For practical reasons, we propose a clear distinction between static data that represent specific completed compilations of data upon which the analyses and conclusions of a given scientific paper may be based, and curated data that belong to a large data collection (usually called a "database") with ongoing goals and curation, for example the various bioinformatics databases that curate ever growing amounts of nucleotide sequences (
Curated data, on the other hand, are usually hosted on external servers or in data hosting centres. A primary goal of the data publishing process in this case is to guarantee that these data are properly described, up to date, available to others under appropriate licensing schemes, peer-reviewed, interoperable, and where appropriate linked from a research article or a data paper at the time of publication. Especially in cases where the long-term viability of the curated project may be insecure (e.g. in the case of grant funded projects) (
Why Publish Data
Data publishing has become increasingly important and already affects the policies of the world's leading science funding frameworks and organizations — see for example the NSF Data Management Plan Requirements, the data management policies of the National Institutes of Health (NIH), Wellcome Trust, or the Riding the Wave (How Europe Can Gain From the Rising Tide of Scientific Data) report submitted to the European Commission in October 2010. More generally, the concept of "open data" is described in the Protocol for Implementing Open Access Data, the Open Knowledge/Data Definition, the Panton Principles for Open Data in Science, and the Open Data Manual. There are several incentives for authors and institutions to publish data (after
- There is a widespread conviction that data produced using public funds should be regarded as a common good, and should be openly published and made available for inspection, interpretation and re-use by third parties.
- Open data increases transparency and the overall quality of research; published datasets can be re-analyzed and verified by others.
- Published data can be cited and re-used in the future, either alone or in association with other data.
- Open data can be integrated with other datasets across both space and time.
- Data integration increases recognition and opportunities for collaboration.
- Open data increases the potential for interdisciplinary research, and for re-use in new contexts not envisaged by the data creator.
- Needless duplication of data-collecting efforts and associated costs will be reduced.
- Published data can be indexed and made discoverable, browsable and searchable through internet services (e.g. Web search engines) or more specific infrastructures (e.g., GBIF for biodiversity data).
- Collection managers can trace usage and citations of digitized data from their collections.
- Data creators, and their institutions and funding agencies, can be credited for their work of data creation and publication through the conventional channels of scholarly citation; priority and authorship is achieved in the same way as with a publication of a research paper.
- Datasets and their metadata, and any related data papers, may be inter-linked into research objects, to expedite and mutually extend their dissemination, to the benefit of the authors, other scientists in their fields, and society at large.
- Published data may be structured as "Linked Data", by which term is meant data accessible using RDF, the Resource Description Framework, one of the fundamentals of the semantic web. Since RDF descriptions are based on publicly available ontology terms, ideally derived from a limited number of complementary ontologies, this permits automated data integration, since data elements from different sources have built-in syntactic and semantic alignment.
How to Publish Data
There are four main routes for scholarly publication of data, most of which are available with various journals and publishers:
- Supplementary files underpinning a research paper and available from the journal's website.
- Data hosted at external repositories but linked back from the research article it underpins.
- Stand-alone description of the data resource in the form of scholarly publication (e.g., Data Paper, or Data Note - see, for example,
Newman and Corke 2009 ,Chavan and Penev 2011 , andCandela et al. 2015 ). - Data published within the article text and downloadable from there in the form of structured data tables or as a result of text mining. This "integrated data publishing" approach has been implemented by the Biodiversity Data Journal (BDJ), which was developed in the course of the EU funded project ViBRANT (
Smith et al. 2013 ). Other examples of a similar approach are executable code published in an article (Veres and Adolfsson 2011 ), or linking of a standard article to an integrated external platform that hosts all data associated with the article, and provides additional data analysis tools and computing resources (an example for that are GigaDB and the GigaScience journal - seeEdmunds et al. 2016 ), or various kinds of implementing 3D visualisations on the basis of MicroCT files (Stoev et al. 2013 ).
Within these main data publishing modes, Pensoft developed a specific set of applications designed to meet the needs of the biodiversity community. Most of these were implemented in the Biodiversity Data Journal and its associated ARPHA Writing Tool (AWT):
- Import of primary biodiversity data from Darwin Core compliant spreadsheets, or manually via a Darwin Core editor, into manuscripts and their consequent publication in a structured and downloadable format (
Smith et al. 2013 ). - Direct online import of Darwin Core compliant primary biodiversity data from GBIF, Barcode of Life, iDigBio, and PlutoF into manuscripts through web services and their consequent publication in a structured and downloadable format (
Senderov et al. 2016 ). - Import of multiple occurrence records of voucher specimens associated with a particular Barcode Index Number (BIN) (
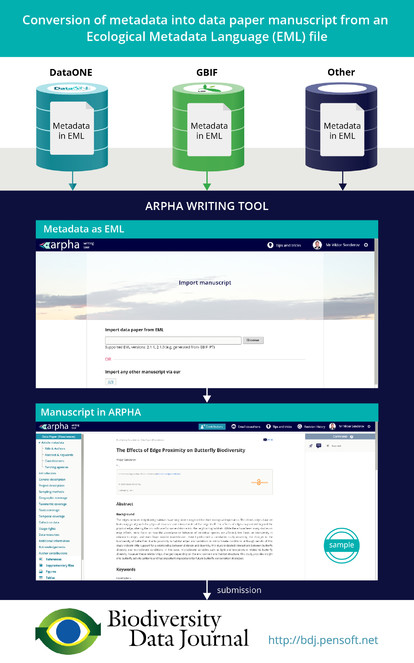
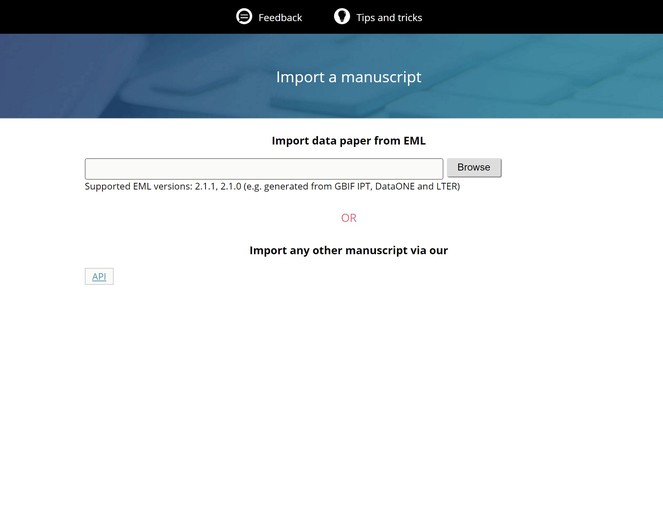
Ratnasingham and Hebert 2013 ) from the Barcode of Life. - Automated generation of data paper manuscripts from Ecological Metadata Language (EML) metadata files stored at GBIF Integrated Publishing Toolkit (GBIF IPT), DataONE, and the Long Term Ecological Research Network (LTER) (
Senderov et al. 2016 , see also Pensoft's blog for details). - Automated export of the occurrence data published in BDJ into Darwin Core Archive (DwC-A) format (
Wieczorek et al. 2012 ) and its consequent ingestion by GBIF. The DwC-A is freely available for download from each article's webpage that contains occurrence data. - Automated export of the taxonomic treatments published in BDJ into Darwin Core Archive. The DwC-A is freely available for download from each article that contains taxonomic treatments data.
- Novel article types in the ARPHA Writing Tool and its associated journals (Biodiversity Data Journal, Research Ideas and Outcomes (RIO Journal), and One Ecosystem): Monitoring Schema, IUCN Red List compliant Species Conservation Profile (
Cardoso et al. 2016 ), IUCN Global Invasive Species Database (GISD) compliant Alien Species Profile, Single-media Publication, Data Management Plan, Research Idea, Grant Proposal, and others. - Nomenclatural acts modelled and developed in BDJ as different types of taxonomic treatments for plant taxonomy.
- Markup and display of biological collection codes against the Global Registry of Biological Repositories (GRBIO) vocabulary (
Schindel et al. 2016 ). - Workflow integration with the GBIF Integrated Publishing Toolkit (IPT) for deposition, publication, and permanent linking between data and articles, of primary biodiversity data (species-by-occurrence records), checklists and their associated metadata (
Chavan and Penev 2011 ). - Workflow integration with the Dryad Data Repository for deposition, publication, and permanent linking between data and articles, of datasets other than primary biodiversity data (e.g., ecological observations, environmental data, genome data and other data types) (see Pensoft blog for details).
- Automated archiving of all articles published in Pensoft's journals in the Biodiversity Literature Respository (BLR) of Zenodo on the day of publication.
Best practice recommendations
- For any form of data publishing, follow the FAIR Data Publishing Principles (
Wilkinson et al. 2016 ). - Follow the Joint Declaration of Data Citation Principles for citation of data in scholarly articles (
Altman et al. 2015 ). - Deposition of data in an established international repository is always to be preferred to supplementary files published on a journal's website.
- Smaller data files, especially those directly underpinning an article, should also be deposited at a data repository and linked from the article. We recommended, however these to be published also as supplementary file(s) to the related article, to ensure an additional joint preservation and presentation of the article together with its associated data.
- If a specialized and well establisdhed repository for a given kind of data exists, it should be preferred over non-specialized ones (see also section "Data Deposition in Open Repositories" below for finer detail), for example:
- Primary biodiversity data (species-by-occurrence) records should be deposited through the GBIF IPT.
- Sample-based biodiversity data (e.g., species abundances from monitoring or inventory studies) should be deposited through the GBIF IPT.
- Genomic data should be deposited at any of the three INSDC repositories (GenBank, European, Nucleotide Archive, ENA and the DNA Databank of Japan, DDBI) either directly or via an affiliated repository, e.g. Barcode of Life Data Systems (BOLD).
- Barcoding and metabarcoding data should be deposited at the Barcode of Life Data Systems (BOLD) or PlutoF.
- Metagenomic data should be deposited at EBI Metagenomics
- Protein sequence data should be deposited at UniProtKB.
- X-ray microtomography (micro-CT) scans should be deposited at Morphosource.
- Phylogenetic data should be deposited at TreeBASE.
- Heterogeneous datasets, or data packages containing various data types should be deposited in generalist repositories, for example Dryad Data Repository, Zenodo, Dataverse, or in another appropriate repository.
- Repositories not mentioned above or in the "Data Deposition in Open Repositories" section below, may be used at the discretion of the author, if they provide long-term preservation of various data types, persistent identifiers to datasets, discoverability, open access to the data, and well proven sustainablility record.
- Digital Object Identifiers (DOIs) or other persistent identifiers (e.g., "stable URIs") to the data deposited in repositories, as well as the name of the repository, should always be published in the paper using or describing that data resource.
- Exceptional cases when publication of data is not possible, or some of the data remain closed or obfuscated, should be discussed with the publisher in advance. In such cases, the authors should provide an open statement explaining why restrictions in open data publishing are needed to be put in force. The author's statement should be published together with the article.
How to Cite Data
This section originates from a draft set of Data Citation Best Practice Guidelines that has been developed for publication by David Shotton, with assistance from colleagues at Dryad and elsewhere, and from earlier papers concerning data citation mechanisms (
The well-established norm for citing genetic data, for example, is that one simply cites the GenBank identifier (accession number) in the text. Similar usage is also commonplace for items in other bioinformatics databases. The latest developments in the implementation of the data citation principles, however, strongly recommend references to data to be included in the reference lists, similarly to literature references (
For such data in data repositories, each published data package and each published data file should always be associated with a persistent unique identifier. A Digital Object Identifier (DOI) issued by DataCite, or CrossRef, should be used wherever possible. If this is not possible, the identifier should be one issued by the data repository or database, and should be in the form of a persistent and resolvable URL. As an example, the use of DOIs in the Dryad Data Repository is explained on the Dryad wiki.
Data citations may relate either to the author's own data, or to data created and published by others ("third-party data"). In the former case, the dataset may have been previously published, or may be published for the first time in association with the article that is now citing it. All these types of data should, for consistency, be cited in the same manner.
Best practice recommendations
As is the norm when citing another research article, any citation of a data publication, including a citation of one's own data, should always have two components:
- An in-text citation statement containing an in-text reference pointer that directs the reader to a formal data reference in the paper's reference list.
- A formal data reference within the article's reference list.
We recommend that the in-text citation statement also contains a separate citation of the research article in which the data were first described, if such an article exists, with its own in-text reference pointer to a formal article reference in the paper's reference list, unless the paper being authored is the one providing that first description of the data. If the in-text citation statement includes the DOI for the data (a strongly desirable practice), this DOI should always be presented as a dereferenceable URI, as shown below. Further to this, both DataCite and CrossRef recommend displaying DOIs within references as full URLs, which serve a similar function as a journal volume, issue and page number do for a printed article, and also give the combined advantages of linked access and the assurance of persistence (
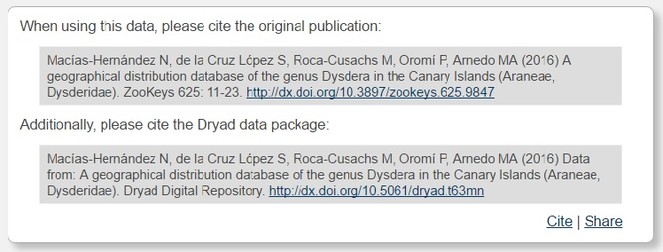
For example, Dryad recommends to cite always both the article in association with which data were published and the data themselves (Fig.
The data reference in the article's reference list should contain the minimal components recommended by the FORCE11 Data Citation Synthesis Group (
- Author(s)
- Year
- Dataset Title
- Data Repository or Archive
- Global Persistent Identifier
- Version, or Subset, and/or Access Date
These components should be presented in whatever format and punctuation style the journal specifies for its references.
The following example demonstrates in general terms what is required.
In-text citation:
“This paper uses data from the [name] data repository at https://doi.org/***** (Jones et al. 2008a), first described in Jones et al. 2008b. “
Data reference and article reference in reference list:
Jones A, Bloggs B, Smith C (2008a). <Title of data package>. <Repository name>. doi: https://doi.org/#####. [Version and/or date of access].
Jones A, Saul D, Smith C (2008b). <Title of journal article>. <Journal> <Volume>: <Pages>. doi: https://doi.org/#####.
Note that the authorship and the title of the data package may, for valid academic reasons, differ from those of the author's paper describing the data: indeed, to avoid confusion of what is being referenced, it is highly desirable that the titles of the data package and of the associated journal article are clearly different.
Requirements for data citation in Pensoft's journals
1. When referring to the author's own newly published data, cited from within the paper in which these data are first described, the citation statement and the data reference should take the following form:
- The citation statement of data deposition should be included in the body of the paper, in a separate section named Data Resources, situated after the Material and Methods section.
- In addition, the formal data reference should be included in the paper's reference list, using the recommended journal's reference format.
The following example demonstrates what is required.
In-text citation:
“The data underpinning the analysis reported in this paper were deposited in the Dryad Data Repository at https://doi.org/10.5061/dryad.t63mn (
AND/OR
"The data underpinning the analysis reported in this paper were deposited in the Global Biodiversity Information Facility (GBIF) at http://ipt.pensoft.net/resource?r=montenegrina&v=1.5 (the URI should be used as identifier only in cases when DOI is not available) (
Data reference in reference list:
Macías-Hernández N, de la Cruz López S, Roca-Cusachs M, Oromí P, Arnedo MA (2016) Data from: A geographical distribution database of the genus Dysdera in the Canary Islands (Araneae, Dysderidae). Dryad Digital Repository. https://doi.org/10.5061/dryad.t63mn [Version and/or date of access].
AND/OR
Feher Z, Szekeres M (2016): Geographic distibution of the rock-dwelling door-snail genus Montenegrina Boettger, 1877 (Mollusca, Gastropoda, Clausiliidae). v1.5. ZooKeys. Dataset/Occurrence deposited in the GBIF. doi: https://doi.org/10.15468/###### OR http://ipt.pensoft.net/resource?r=montenegrina&v=1.5, (the latter to be used in cases when DOI is not available). [Version and/or date of access].
2. When acknowledging re-use in the paper of previously published data (including the author's own data) that is associated with another published journal article, the citation and reference should take the same form, except that the full correct DOI should be employed, and that the journal article first describing the data should also be cited:
- A statement of usage of the previously published data, with citation of the data source(s) and of the related journal article(s), should be placed in a separate section named Data Resources, situated after the Material and Methods section.
- In addition, the formal data reference and a formal reference to the related journal article should be included in the paper's reference list, using the recommended journal's reference format.
The following example demonstrates what is required.
In-text citation:
“The data underpinning this analysis were obtained from the Dryad Data Repository at https://doi.org/10.5061/dryad.t63mn (
Data reference and article reference in reference list:
Macías-Hernández N, de la Cruz López S, Roca-Cusachs M, Oromí P, Arnedo MA (2016) A geographical distribution database of the genus Dysdera in the Canary Islands (Araneae, Dysderidae). ZooKeys 625: 11-23. https://doi.org/10.3897/zookeys.625.9847.
Macías-Hernández N, de la Cruz López S, Roca-Cusachs M, Oromí P, Arnedo MA (2016) Data from: A geographical distribution database of the genus Dysdera in the Canary Islands (Araneae, Dysderidae). Dryad Digital Repository. https://doi.org/10.5061/dryad.t63mn [Version and/or date of access].
3. When acknowledging re-use of previously published data (including the author's own data) that has NO association with a published research article, the same general format should be adopted, although a reference to a related journal article clearly cannot be included:
- A statement of usage of previously published data, with citation of the data source(s), should be placed in a separate section named Data Resources, situated after the Material and Methods section.
- In addition, the formal data reference should be included in the paper's reference list, using the recommended journal's reference format for data citation.
The following real example demonstrates what is required.
In-text citation:
“The present paper used data deposited by the Zoological Institute of the Russian Academy of Sciences in the Global Biodiversity Information Facility (GBIF) at https://doi.org/10.15468/c3eork (
Data reference in reference list:
Volkobitsh M, Glikov A, Khalikov R (2017) Catalogue of the type specimens of Polycestinae (Coleoptera: Buprestidae) from research collections of the Zoological Institute, Russian Academy of Sciences. Zoological Institute, Russian Academy of Sciences, St. Petersburg, deposited in GBIF. https://doi.org/10.15468/C3EORK. [Version and/or date of access].
Data Publishing Policies
General Policies for Biodiversity data
One of the basic postulates of the Panton Principles is that data publishers should define clearly the license or waiver under which the data are published, so re-use rights are clear to potential users. They recommend use of the most liberal licenses, or of public domain waivers, to prevent legal and operational barriers for data sharing and integration. For clarity, we list here the short version of the Panton Principles:
- When publishing data, make an explicit and robust statement of your wishes regarding re-use.
- Use a recognized waiver or open publication license that is appropriate for data.
- If you want your data to be effectively used and added to by others, it should be fully "open" as defined by the Open Knowledge/Data Definition – in particular, non-commercial and other restrictive clauses should not be used.
- Explicit dedication of data underlying published science into the public domain via PDDL or CC-Zero is strongly recommended and ensures compliance with both the Science Commons Protocol for Implementing Open Access Data and the Open Knowledge/Data Definition.
A domain-specific implementation of the open access principles for biodiversity data was elaborated during the EU project pro-iBiosphere and resulted in the widely endorsed Bouchout Declaration for Open Biodiversity Knowledge Management. Further, the EU project EU BON analysed the current copyright legislation and data policies in various European countries and elaborated a set of best practice recommendations (
- Promoting the understanding that primary biodiversity data are facts and therefore NOT a subject of copyright; they belong to the public domain, independent of their source;
- Requiring explicit statements that clearly place biodiversity data in the public domain, by applying a standardised waiver for any eventual copyright or database protection right, for example Creative Commons Zero (CC0). Some countries may still need special licenses for data irrespective of its source (cf. https://github.com/unitedstates/licensing/issues/31).
- To the maximum possible extent, rendering printed materials, PDFs, and other non-machine-actionable biodiversity data and narratives, into machine-readable and harvestable formats.
Data Publishing Licenses
In practice, a variety of waivers and licenses exist that are specifically designed for and appropriate for the treatment of data, as listed in Table
Data publishing licenses recommended by Pensoft.
| Data publishing license | URL |
| Open Data Commons Attribution License | http://www.opendatacommons.org/licenses/by/1.0/ |
| Creative Commons CC-Zero Waiver | http://creativecommons.org/publicdomain/zero/1.0/ |
| Open Data Commons Public Domain Dedication and License | http://www.opendatacommons.org/licenses/pddl/1-0/ |
The default data publishing license used by Pensoft is the Open Data Commons Attribution License (ODC-By), which is a license agreement intended to allow users to freely share, modify, and use the published data(base), provided that the data creators are attributed (cited or acknowledged).
As an alternative, the other licenses or waivers, namely the Creative Commons CC0 waiver (also cited as “CC-Zero” or “CC-zero”) and the Open Data Commons Public Domain Dedication and Licence (PDDL), are also STRONGLY encouraged for use in Pensoft journals. According to the CC0 waiver, "the person who associated a work with this deed has dedicated the work to the public domain by waiving all of his or her rights to the work worldwide under copyright law, including all related and neighbouring rights, to the extent allowed by law. You can copy, modify, distribute and perform the work, even for commercial purposes, all without asking permission."
Publication of data under a waiver such as CC0 avoids potential problems of "attribution stacking" when data from several (or possibly many) sources are aggregated, remixed or otherwise re-used, particularly if this re-use is undertaken automatically. In such cases, while there is no legal requirement to provide attribution to the data creators, the norms of academic citation best practice for fair use still apply, and those who re-use the data should reference the data source, as they would reference others' research articles.
The Attribution-ShareAlike Open Data Commons Open Database License (OdbL) is NOT recommended for use in Pensoft's journals, because it is very difficult to comply with the share-alike requirement in scholarly publishing. Nonetheless, it may be used as an exception in particular cases.
Many widely recognized open access licenses are intended for text-based publications to which copyright applies, and are not intended for, and are not appropriate for, data or collections of data which do not carry copyright. Creative Commons licenses apart from CC-Zero waiver (e.g., CC-BY, CC-BY-NC, CC-BY-NC-SA, CC-BY-SA, etc.) as well as GFDL, GPL, BSD and similar licenses widely used for open source software, are NOT appropriate for data, and their use for data associated with Pensoft journal articles is strongly discouraged.
Authors should explicitly inform the publisher if they want to publish data associated with a Pensoft journal article under a license that is different from the Open Data Commons Attribution License (ODC-By), Creative Commons CC0, or Open Data Commons Public Domain Dedication and Licence (PDDL).
Any set of data published by Pensoft, or associated with a journal article published by Pensoft, must always clearly state its licensing terms in both a human-readable and a machine-readable manner.
Where data are published by a public data repository under a particular license, and subsequently associated with a Pensoft research article or data paper, Pensoft journals will accept that repository license as the default for the published datasets.
Images, videos and similar "artistic works" are usually covered by copyright "automatically", unless specifically placed in the public domain by use of a public domain waiver such as CC0. Where copyright is retained by the creator, such multimedia entities can still be published under an open data attribution license, while their metadata can be published under a CC0 waiver.
Databases can contain a wide variety of types of content (images, audiovisual material, and sounds, for example, as well as tabular data, which might all be in the same database), and each may have a different license, which must be separately specified in the content metadata. Databases may also automatically accrue their own rights, such as the European Union Database Right, although no equivalent database right exists in the USA. In addition, the contents of a database, or the database itself, can be covered by other rights not addressed here (such as private contracts, trademark over the name, or privacy rights / data protection rights over information in the contents). Thus, authors are advised to be aware of potential problems for data re-use from databases, and to clear other rights before engaging in activities not covered by the respective license.
Data Deposition in Open Repositories
General Information
Open data repositories (public databases, data warehouses, data hosting centres) are subject- or institution-oriented infrastructures, usually based at large national or international institutions. These provide data storage and preservation according to widely accepted standards, and provide free access to their data holdings for anyone to use and re-use under the minimum requirement of attribution, or under an open data waiver such as the CC0 waiver. We do NOT include here and do NOT recommend for use repositories which provide data after permission or by other methods of human-controlled registration.
Advantages of depositing data in internationally recognised repositories include:
- Visibility: Making your data available online (and linking it to the publication) provides an independent way for others to discover your work.
- Citability: all data you deposit will receive a persistent, resolvable identifier that can be used in a citation, as well as listed on your CV.
- Workload reduction: if you receive individual requests for data, you can simply direct them to the files in Dryad.
- Preservation: your data files will be safely archived in perpetuity.
- Impact: other researchers have more opportunities to use and cite your work.
There are several directories of data repositories relevant to biodiversity and ecological data, such as re3data, or those listed in the Open Access Directory, or in Table 2 of
A very useful resource that puts together information on journal data policies, repositories, and standards grouped by domain, type of data, and organisation is BioSharing (
Such repositories could be used to host data associated with a published data paper, as explained below. For their own data, authors are advised to use an internationally recognised, trusted (normally ISO-certified), specialized repository (see
- PANGAEA. The information system PANGAEA operates as an open data repository aimed at archiving, publishing and distributing georeferenced data from earth system research. Each dataset can be identified and cited by using a DOI. Data are archived as supplements to publications or as citable data collections. Data citations are available through the portal of the German National Library of Science and Technology (GetInfo). Data management and archiving policies follow the recommendations of the Commission on Professional Self Regulation in Science, the ICSU Data Sharing Principles, and the OECD Principles and Guidelines for Access to Research Data from Public Funding. PANGAEA is open to any project or individual scientist to archive and publish data. Data submission can be started here.
- The Knowledge Network for Biocomplexity (KNB) is a USA-based national network intended to facilitate data management and preservation in ecological and environmental research. For scientists, the KNB is an efficient way to discover, access, interpret, integrate and analyze complex ecological data from a highly-distributed set of field stations, laboratories, research sites, and individual researchers.
- DataBasin is a free system that connects the users with spatial datasets, non-technical tools, and a network of scientists and practitioners. One can explore and download a vast library of datasets, upload and publish own data, create working groups, and produce customized maps that can be easily shared.
- DataONE provides the distributed framework, management, and robust technologies that enable long-term preservation of diverse multi-scale, multi-discipline, and multi-national observational data. DataONE initially emphasises observational data collected by biological (genome to ecosystem) and environmental (atmospheric, ecological, hydrological, and oceanographic) scientists, research networks, and environmental observatories.
- Dataverse is an open source web application to share, preserve, cite, explore, and analyze research data. A Dataverse repository is the software installation, which then hosts multiple dataverses. Each dataverse contains datasets, and each dataset contains descriptive metadata and data files (including documentation and code that accompany the data). As an organising method, dataverses may also contain other dataverses. Some dataverses may have non-CC0 data, for example the Singaporean NTU dataverse has CC-BY-NC licensed data by default.
- Protocols.io is an open access platform for publishing, sharing and finding life science research protocols.
Taxonomy
There are several aggregators and registries of taxonomic data, which differ in their content, policies and methods of data submisison.
- Catalogue of Life (CoL) is a comprehensive and authoritative global index of species and their associated taxonomic hierarchy. The Catalogue holds essential information on the names, relationships and distributions of over 1.6 million species, continuosly compiled from 158 contributing databases around the world (as of February 2017). The Catalogue of Life is led by Species 2000, working in partnership with the Integrated Taxonomic Information System (ITIS). Authors can submit their global taxon checklists to the CoL editors following a template and guidelines.
- Integrated Taxonomic Information System (ITIS) is established by several federal agencies of the USA to provide an authoritative taxonomic information on species of plants, animals, fungi, and microbes, and their hierarchical classification, with a focus on North America. Potential contributors to ITIS are encouraged to view the ITIS Submittal Guidelines.
- NCBI Taxonomy serves as a taxonomic backbone for the National Center for Biotechnology Infomation of the USA (NCBI), and its services, for example GenBank. NCBI Taxonomy is not a single taxonoimic treatise, but rather a compiler of taxonomic information from a variety of sources, including the published literature, web-based databases, and the advice of sequence submitters and outside taxonomy experts.
-
International Plant Name Index (IPNI) is a collaborative effort between The Royal Botanic Gardens, Kew, the Harward University Herbaria, and the Australian National Herbarium to provide a single point of registration and reference for the names and associated basic bibliographical details of seed plants, ferns and lycophytes. The data are gathered and curated by a team of editors from the published literature and are freely available to the community. The pre-publication indexing of new plant taxa and nomenclatural acts in IPNI and inclusion of the IPNI identifiers in the original descriptions (protologues) was first trialled and made a routine practice in the journal PhytoKeys since the publication of its first issue in 2011 (
Kress and Penev 2011 ,Knapp et al. 2011 ). Later, Pensoft created an automated registration pipeline for new names in IPNI (Penev et al. 2016 ). -
MycoBank is the leading online database, established by the International Mycological Association (IMA), for documenting new names and combinations of fungy, and associated data, for example descriptions and illustrations. The nomenclatural novelties are assigned a unique MycoBank identifier that can be cited in the publication where the nomenclatural novelty is introduced. These identifiers are also used by the nomenclatural database Index Fungorum. As a result of changes to the International Code for Nomenclature of algae, fungi, and plants, ICNafp) (previously International Code for Botanical Nomenclature, ICBN), pre-publication registration of names and inclusion of record identifiers in the published protologues is mandatory since January 1st 2013 (see
Hawksworth 2011 ). - Index Fungorum is a global fungal nomenclator currently coordinated and supported by The Royal Botanic Gardens, Kew. Index Fungorum contains names of fungi (including yeasts, lichens, chromistan fungal analogues, protozoan fungal analogues and fossil forms) at all ranks. Index Fungorum now provides a mechanism to register names of new taxa, new names, new combinations and new typifications following the changes to the ICNafp (see above).
-
ZooBank is the Official Register of the International Commission on Zoological Nomenclature (ICZN) for registration of new nomenclatural acts, published works, and authors. Since 1st of January 2012, pre-publication registration in ZooBank has become mandatory for electronic-only publications (
International Commission on Zoological Nomenclature 2012 ). The Pensoft journal ZooKeys was the first to apply a mandatory registration of new zoological names at ZooBank since the publication of its first issue in 2008 (Penev et al. 2008 ). Authors publishing nomenclatural novelties in Pensoft journals do not have to deal with registration of these at ZooBank because it is provided in-house, through an automated pipeline (Penev et al. 2016 ). - PaleoBiology Database is a public resource whose purpose is to provide global, collection-based occurrence and taxonomic data for marine and terrestrial animals and plants of any geological age, as well as web-based software for statistical analysis of the data. The project's wider, long-term goal is to encourage collaborative efforts to answer large-scale paleobiological questions by developing a useful database infrastructure and bringing together large data sets. There is an option to protect data for private use only.
-
TreatmentBank is a resource that stores and provides access to taxonomic treatements and data therein, extracted from the literature. TreatmentBank is established by Plazi, who also provide a tool for text mining and data extraction called GoldenGATE Document Editor. Authors who publish in Pensoft's journals do not have to deal with deposition of their taxonomic treatments to Plazi, as the latter are harvested automatically using the TaxPub extension to the Journal Archival Tag Suite (JATS) (
Catapano 2010 ,Penev et al. 2012 ).
Species-by-Occurrence and Sample-Based data
The Global Biodiversity Information Facility (GBIF) was established in 2001 and is now the world's largest multilateral initiative for enabling free and open access to biodiversity data via the Internet. It comprises a network of 54 countries and 39 international organisations that contribute to its vision of "a world in which biodiversity information is freely and universally available for science, society, and a sustainable future". It seeks to fulfil this mission by promoting an international data infrastructure through which institutions can publish data according to common standards, thus enabling research that had not been possible before. The GBIF network facilitates access to over 704 million species occurrences in 30,894 datasets sourced from 867 data-publishing institutions (as of January 2017).
GBIF is not a repository in the strict sense, but a distributed network of data publishers and local data hosting centres that publish data based on community-agreed standards for exchange/sharing of primary biodiversity data. At a global scale, discovery and access to data is facilitated through the GBIF data portal. Pensoft facilitates publishing of data and metadata to the GBIF network through Pensoft’s IPT Data Hosting Center, which is based on the GBIF Integrated Publishing Toolkit (IPT) (
The Darwin Core Archive (DwC-A) (see also http://rs.tdwg.org/dwc/terms/guides/text/index.htm and
- One or more data files keeping all records of the particular dataset in a tabular format such as a comma-separated or tab-separated list;
- The archive descriptor (meta.xml) file describing the individual data file columns used, as well as their mapping to DwC terms; and
- A metadata file describing the entire dataset which GBIF recommends to be based on EML (Ecological Metadata Language 2.1.1).
The format is defined in the Darwin Core Text Guidelines. Darwin Core is no longer restricted to occurrence data, and together with the more generic Dublin Core metadata standard (on which its ideas are based), it is used by GBIF and others to encode data about organism names, taxonomies, species information, and, more recently, sample data (i.e., data from ecological/environmental investigations that are typically quantitative and adhere to standardised protocols, so that changes and trends in populations can be detected).
GBIF has produced a series of documents and supporting tools that focus primarily on data publishing using the Darwin Core standard. Guides are available for publishing:
Besides the GBIF Integrated Publishing Toolkit, there are two additional tools developed for producing Darwin Core Archives:
- The Darwin Core Archive Spreadsheet Processor provides a set of MS Excel templates, which are coupled with a web service that processes completed files and returns a validated Darwin Core Archive. Templates exist for primary biodiversity data, simple checklists, and EML metadata. See http://tools.gbif.org/spreadsheet-processor/ for further details.
- The Darwin Core Archive Assistant is a browser-based tool that composes an XML metafile, the only XML component of a Darwin Core Archive. It displays a drop-down list of Darwin Core and extension terms, accessed dynamically from the GBIF registry, and displays these to the user who describes the data files. This allows Darwin Core Archives to be created for sharing without the need to install any software. See: http://tools.gbif.org/dwca-assistant/ for details.
The Darwin Core Archive (DwC-A) files can be used to publish data underlying any taxonomic revision or checklist through the GBIF IPT or as supplementary files (see
As of version 2.2, the GBIF IPT incorporates use of DOIs allowing data publishers to automatically connect with either DataCite or EZID for DOI assignment. GBIF will issue DOIs for all newly published datasets where absent while recognizing and displaying publisher-assigned DOIs for existing datasets. The GBIF IPT now also requires publishers to select one of three standardised machine-readable data waivers or licenses (CC0, CC-BY, CC-BY-NC) for their data to clarify the conditions for re-use.
Images
Images can be deposited at generic repositories, such as Zenodo, figshare or Flickr. There are also specialized repositories for biodiversity images:
- Morphbank is a database of images and metadata used for international collaboration, research and education. Images deposited in Morphbank :: Biological Imaging document a wide variety of research including: specimen-based research in comparative anatomy, morphological phylogenetics, taxonomy and other biodiversity-related fields.
- Morphosource is a project-based data archive that allows researchers to store and organize, share, and distribute 3D data, for example raw microCt data and surface meshes representing vouchered specimens. File formats include tiff, dicom, stanford ply, and stl.
- Biodiversity Literature Repository (BLR) at Zenodo (see details in the section Biodiversity Literature below).
Phylogenies
There are relatively few repositories dealing with phylogentic data, of which we recommend the following:
-
TreeBASE is a repository of phylogenetic information, specifically user-submitted phylogenetic trees and the data used to generate them. TreeBASE accepts all types of phylogenetic data (e.g., trees of species, trees of populations, trees of genes) representing all biotic taxa. Data in TreeBASE are exposed to the public if they are used in a publication that is in press or published in a peer-reviewed scientific journal, book, conference proceedings, or thesis. Data used in publications that are in preparation or in review can be submitted to TreeBASE but are embargoed until after publication, and only available before publication to the publication editors or reviewers using a special access code. We also recommend following the best practices for sharing and publishing phylogenies, as detailed in
Stoltzfus et al. (2012) andCranston et al. (2014) . - MorphoBank is an online database and workspace for images and affiliate data with those images (labels, species names, etc.) used in evolutionary research in systematics. MorphoBank provides a platform for live collaboration on phylоgenetic matrices by teams in a private workspace where they can interlink images with phylogenetic matrices. MorphoBank stores: phylogenetic matrices (Nexus or TNT format), 2D (including JPEG, GIF, PNG, TIFF and Photoshop) and 3D (PLY, STL, ZIP, TIFF and DCM) image data and video (MPEG-4, QuickTime and WindowsMedia). MorphoBank also offers a Documents folder for additional files related to the research, such as PDFs, Word documents, and text files (e.g., morphometric data, phylogenetic trees).
Gene Sequence
Pensoft journals collaborate with four repositories for genomic data, albeit with the assumption that no matter where gene sequence data will be deposited, they should finally be submitted also to GenBank. Data and metadata formatting should comply with the Genomic Standards Consortium (GSC) sample metadata guidelines respectively, allowing data interoperability across the wider genomics community. Inclusion of the hyperlinked accession numbers in the article is a prerequisite for publication in Pensoft journals. The most important repositories for genomic data are:
- International Nucleotide Sequence Database Collaboration (INSDC), mostly known through its founding partner, the NCBI GenBank. GenBank is a genetic sequence database, an annotated collection of all publicly available DNA sequences. Hosted at the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM) under the umbrella of the National Institutes of Health (NIH), GenBank is part of the International Nucleotide Sequence Database Collaboration (INSDC), which also comprises the DNA DataBank of Japan (DDBJ) and the European Bioinformatics Institute (EBI), which is part of the European Molecular Biology Laboratory (EMBL-EBI). Raw sequencing data in particular needs to be deposited in one of the INSDC data repositories, such as the NCBI Sequence Read Archive (SRA), the EBI European Nucleotide Archive (ENA), or the DDBJ Sequence Read Archive (DRA). These three organizations exchange data on a daily basis. They also handle assembled and annotated sequence data, as well as host a number of linked databases that handle more processed data types like variation data (with both the EBI and NCBI handling genetic and structural variants). An example of a GenBank record for a Saccharomyces cerevisiae gene may be viewed here. There are several options for submitting data to GenBank.
- The Barcode of Life Data System (BOLD) is established at the University of Guelph as "an informatics workbench aiding the acquisition, storage, analysis, and publication of DNA barcode (mostly COI) records" (
Ratnasingham and Hebert 2007 ). BOLD have an agreement with GenBank for deposition of barcode (COI) sequences in GenBank as well through a web-based Barcode Submission Tool. - PlutoF Biodiversity Platform has been developed originally as a data management platform for barcode data based mostly on the Internal Transcribed Spacer (ITS) region as most suitable for the identification of fungi. Recently PlutoF was developed into a platform to "create, manage, share, analyse and publish biology-related databases and projects". PlutoF have an agreement with GenBank for deposition of gene sequences in GenBank.
Best practice recommendations for biodiversity genomic data
- Always aim at depositing data before submission of the manuscript, so that they can be linked to and from the manuscript and are made freely available for peer-review. Even if not yet public during the review process, reviewer access is available via NCBI.
- Gene sequences should always be published in GenBank, either directly or through INSDC, even if they are openly available in other repositories.
- A paper dealing with gene sequences should always contain the GenBank accession numbers, and where possible should use the BioProject accession (formatted PRJxxxxx) as well.
- When including gene sequences deposited in other repositories, authors should provide hyperlinked identifiers (e.g. accession numbers) of those records in the manuscript text.
- It is strongly recommended to publish large genomic databases, or separate species genomes, or barcode reference libraries in the form of data papers or "BARCODE data release papers". A BARCODE data release paper is a short manuscript that announces and documents the public deposit to a member of the INSDC of a significant body of data records that meets the BARCODE data standards (for examples, see
Rougerie et al. 2015 ,Schindel et al. 2011 ).
Protein Sequence
- The Protein Data Bank (PDB) created by the the Research Collaboratory for Structural Bioinformatics (RCSB) contains information about experimentally-determined structures of proteins, nucleic acids, and complex assemblies. As a memberof the Worldwide Protein Data Bank (wwPDB), the RCSB PDB curates and annotates PDB data according to agreed standards. See its data deposition policies and services.
- The Universal Protein Resource (UniProt) is a comprehensive resource for protein sequence and annotation data. The UniProt databases are the UniProt Knowledgebase (UniProtKB), the UniProt Reference Clusters (UniRef), and the UniProt Archive (UniParc).
Genomics
- ArrayExpress (EMBL-EBI) and the NCBI Gene Expression Omnibus (NCBI GEO) are archives for data from high-throughput functional genomics experiments such as microarrays and sequencing based approaches such as RNA-seq, miRNA-seq, ChIP-seq, methyl-seq, etc. Data are collected to MIAME (Minimum Information About a Microarray Experiment) and MINSEQE (Minimum Information about a high-throughput SEQuencing Experiment) standards. Experiments are submitted directly to ArrayExpress or are imported from the NCBI GEO database and vice versa. ArrayExpress and NCBI GEO are strongly recommended for deposition of species genomes or transcriptomes, metagenomic and other biodiversity-related functional genomics data. For high-throughput sequencing based experiments the raw data is brokered to the EBI or GenBank, while the experiment descriptions and processed data are archived in these databases.
- EBI Metagenomics (EMBL-EBI) is a pipeline for the analysis and archiving of metagenomic data automatically archived in the European Nucleotide Archive (ENA) and intended for public release.
-
GigaDB primarily serves as a repository to host data and tools associated with articles in the journal GigaScience; however, it also includes a subset of datasets that are not associated with GigaScience articles. GigaDB defines a dataset as a group of files (e.g., sequencing data, analyses, imaging files, software programs) that are related to and support an article or study. An example of multifunctional use of GigaDB or various types of biodiversity data is the paper of
Stoev et al. (2013) and its associated editorial (Edmunds et al. 2013 ).
Other Omics
Metabolomics
Metabolomics data should be deposited in any of the member databases of the Metabolomexchange data aggregation and notification consortium. Such partners, for example, are the EMBL-EBI MetaboLights repository and the Metabolomics Workbench of NIH, which are data archives for metabolomics experiments and derived information.
Proteomics
Proteomics data should be deposited in any of the members of the ProteomeXchange consortium and following the MIAPE (The Minimum Information About a Proteomics Experiment) guidelines. The founding members of ProteomeXchange are Pride, the PRoteomics IDEntifications Database at the EMBL-EBI and PeptideAtlas, part of the Institute of Systems Biology in Seattle, USA. The other two repositories at ProteomeXchange are MassIVE and jPost.
Various Data Types
Dryad Data Repository
Pensoft encourages authors to deposit data underlying biological research articles in the Dryad Data Repository in cases where no suitable more specialized public data repository (e.g., GBIF for species-by-occurrence data and taxon checklists, or GenBank for genome data) exists. Dryad is particularly suitable for depositing data packages consisting of different types of data, for example datasets of species occurrences, environmental measurements, and others.
Pensoft supports Dryad and its goal of enabling authors to publicly archive sufficient data to support the findings described in their journal articles. Dryad is a safe, sustainable location for data storage, and there are no restrictions on data format. Note that data deposited in Dryad are made available for re-use through the Creative Commons CC0 waiver, detailed above.
Data deposition in Dryad is a subject to a small charge that the authors or their institutions should regulate directly with Dryad.
Data can be deposited with Dryad either before or at the time of submission of the manuscript to the journal, or after the manuscript acceptance but before submission of the finally revised, ready-for-layout version for publication. Nonetheless, the authors should always aim at depositing data before submission of the manuscript, so that they can be linked both from and to the manuscript and made freely available for peer-review.
The data deposition at Dryad is integrated with the workflow in Pensoft's ARPHA Journal Publishing System. The acceptance letters automatically generated by email by Pensoft's journals on the day of acceptance of a manuscript contain instructions on how to upload data underpinning the article to Dryad, if desired by the authors (see this blog post for details).
Once you deposit your data package, it receives a unique and stable identifier, namely a DataCite DOI. Individual data files within this package are given their own DOIs, based on the package DOI, as do subsequent versions of these data files, as explained under DOI usage on the Dryad wiki. You should include appropriate Dryad DOIs in the final text of the manuscript, both in the in-text citation statement in the Data Resources section and in the formal data reference in your paper's reference list, as explained and exemplified above. This is very important, since if the data DOI does not appear in the final published article, that greatly weakens its connection to the underlying data.
More information about depositing data in Dryad can be found at http://www.datadryad.org/repo/depositing.
You may wish to take a look at some example data packages in Dryad to see how data packages related to published articles are displayed, such as doi: 10.5061/dryad.7994 and doi: 10.5061/dryad.8682.
Data deposited in Dryad in association with Pensoft journal articles will be made public immediately upon publication of the article.
Zenodo
Zenodo is a research data repository launched in 2013 by the EU-Funded OpenAIRE project and CERN to provide a place for researchers to deposit datasets of up to 50 GB in any subject area. Zenodo code is open source, and is built on the foundation of the Invenio digital library which is also open source. The work-in-progress, open issues, and roadmap are shared openly in GitHub, and contributions to any aspect are welcomed from anyone. All metadata is openly available under CC0 waiver, and all open content is openly accessible through open APIs.
Zenodo assigns a DataCite DOI to each stored research object, or uses the original DOIs of the articles or research objects, if available. Scientists may use Zenodo to store any kind of data that can thereafter be linked to and cited in research articles.
The repository allows non-open-access materials to be uploaded but not displayed in public, except for their metadata which are freely available under the CC0 waiver.
Biodiversity Literature
Biodiversity Heritage Library (BHL) is a searchable archive of scanned public domian books and journals. Originally BHL was focusing mostly on the historical biodiversity literature, however now it is possible to incorporate also materials that are still under copyright through agreements with publishers. Pensoft journals harvest the BHL content for mentions of taxon names and display the original sources through the Pensoft Taxon Profile tool. Bibliographical metadata of the articles published in Pensoft's journals are submitted to BHL on the day of publication. On the top of the BHL content, Roderick Page from the University of Glasgow built BioStor as an open source application that searches and displays the BHL articles by article metadata and individual pages.
The Biodiversity Literature Repository (BLR) is an open community repository at Zenodo built by Plazi and Pensoft to archive articles, images and data in the biodiversity domain. Plazi uploads article PDFs and other materials extracted from legacy literature through their GoldenGATE Imagine tool. Pensoft journals are automatically archiving in BLR all biodiversity-related articles, supplementary files and individual images, through Web services, on the day of publication. The uploaded materials are archived at Zenodo under their own DOIs, if exisiting, or are assigned Zenodo DOIs.
The Bibliography of Life (BoL) was created by the EU FP7 project ViBRANT to search, retrieve and store bibliographic references and is currently maitained by Pensoft and Plazi. BoL consists of the search and discovery tool ReFindit and a repository for bibliographic references harvested from the literature, RefBank.
Guidelines for Authors
Data Published within Supplementary Information Files
Online publishing allows an author to provide data sets, tables, video files, or other information as supplementary information files associated with papers, or to deposit such files in one of the repositories described above, which can greatly increase the impact of the submission. For larger biodiversity datasets, authors should consider the alternative of submitting a separate data paper (see description below).
Submission of data to a recognised data repository is encouraged as a superior and more sustainable method of data publication than submission as a supplementary information file with an article. Nevertheless, Pensoft will accept supplementary information files if authors wish to submit them with their articles and demonstrate that no suitable repository exists. Details for uploading such files are given in Step 4 of the Pensoft submission process (example from ZooKeys) available through the “Submit a Manuscript” button on any of the Pensoft journal websites.
By default, the maximum file size for each supplementary information file that can be uploaded onto the Pensoft web site is 50 MB. If you need more than that, or wish to submit a file type not listed below, please contact Pensoft's editorial office before uploading.
When submitting a supplementary information file, the following information should be completed:
- File format (including name and a URL of an appropriate viewer if the format is unusual).
- Title of the supplementary information file (the authorship will be assumed to be the same as for the paper itself, unless explicitly stated otherwise).
- Description of the data, software listings, protocols or other information contained within the supplementary information file.
All supplementary information files should be referenced explicitly by file name within the body of the article, e.g. “See Supplementary File 1: Movie 1 for a recording of the original data used to perform this analysis”.
The ARPHA Writing Tool and the journals currently based on it (Biodiversity Data Journal, Research Ideas and Outcomes, One Ecosystem, and BioDiscovery) provide the functionality to cite the supplementary materials through in-text citations in the same way as figures, tables or references are cited.
Ideally, the supplementary information file formats should not be platform specific, and should be viewable using free or widely available tools. Suitable file formats are:
For supplementary documentation:
- RTF (Rich Text Format)
- PDF (Adobe Acrobat; ISO 32000-1)
- HTML (Hypertext Markup Language)
- XML (Extensible Markup Language)
For animations:
- SWF (Shockwave Flash)
- DHTML (Dynamic HTML)/HTML5
For images:
- SVG (Scalable Vector Graphics)
- GIF (Graphics Interchange Format)
- JPEG/JFIF (JPEG File Interchange Format)
- PNG (Portable Network Graphics)
- TIFF (Tagged Image File Format)
For movies:
- MOV (QuickTime)
- MPG (MPEG)
- OGG (an open and free multimedia container format)
- WebM (an open and free multimedia container format)
For datasets:
- CSV (Comma separated values)
- TSV (Tab separated values)
The file names should use the standard file extensions (as in “Supplementary-Figure-1.png”). Please also make sure that each supplementary information file contains one particular data type, or is of a single table, figure, image or video.
To facilitate comparisons between different pieces of evidence, it is common to produce composite figures or to concatenate originally separate recordings into a single audio or video file. We do not recommend such practice, since it is often simpler to just open the two (or more) raw files in question and to appreciate and manipulate them side by side, and such concatenation is a barrier to re-use. Likewise, we do not recommend to provide metadata in non-editable ways (e.g., adding a letter or an arrow into bitmap images or video frames), which complicates re-use too (e.g. translation into another language, or zooming in for additional details).
Best practice recommendations
- Open data formats should be preferred over proprietary ones (for example, for spreadsheets, CSV should always be preferred over XLS).
- Always follow community-accepted standards within the respective scientific domain (if such exist) when formatting data files, because this will make your data interoperable with other data in the same domain.
- To maximise interoperability, plain-text data files should be UTF-8 encoded with no embedded line breaks.
- For species-by-occurrence data, the authors are strongly encouraged to publish these through the GBIF Integrated Publishing Tookit (see above) first, then link to the data in the "Data resources" section of the article and also cite the dataset in the reference section via its GBIF DOI or the GBIF IPT unique HTTP identifier. In addition, authors may also publish the same data as supplementary files to the article in Darwin Core Archive. The Darwin Core Archive of the data can be downloaded from the GBIF IPT or created in another way.
- For species-by-occurrence data published as supplementary files to the article, authors should use a Darwin Core compliant spreadsheets or tabular text files (http://arpha.pensoft.net/lib/files/Species_occurrence-1_v1_DwC_Template.xls).
Import of Darwin Core Specimen Records into Manuscripts
This specific functionality is available in the ARPHA Writing Tool (AWT) and currently being used in the "Materials" subsection of the "Taxon treatment" section in the "Taxonomic paper" template of the Biodiversity Data Journal. Darwin Core compliant specimen records can be imported into structured format in the manuscript text in three ways:
- manually through the Darwin Core compliant HTML editor embedded in the AWT,
- from a Darwin Core compliant spreadsheet template (for example, from an Excel spreadsheet; the template is available in the AWT through the link http://arpha.pensoft.net/lib/files/Species_occurrence-1_v1_DwC_Template.xls),
- automatically, through web services from online biodiversity data platforms (GBIF, Barcode of Life, iDigBio, and PlutoF).
While the first two methods of data import speak for themselves and one could easily implement them following the instructions on the user interface, the third one deserves a more detailed description, as it is still unique in the data publishing landscape.
The workflow has been thoroughly described from the user's perspective in a blog post and in the paper of
- At one of the supported data portals (GBIF, Barcode of Life, iDigBio, and PlutoF), the author locates the specimen record he/she wants to import into the Materials section of a Taxon treatment (available in the Taxonomic Paper manuscript template in the Biodiversity Data Journal).
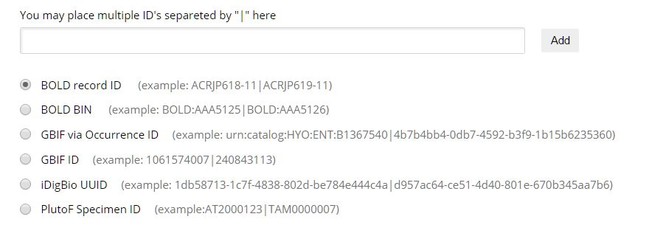
- Depending on the portal, the user finds either the occurrence identfier of the specimen, or a database record identifier of the specimen record, and copies that into the respective upload field of the ARPHA system (Fig.
3 ). - After the user clicks on "Add," a progress bar is displayed, while the specimens are being uploaded as material citations.
- The new material citations are rendered in both human- and machine-readable DwC format in the Materials section of the respective Taxon treatment and can be further edited in AWT, or downloaded from there as a CSV file.
Data Published in Data Papers
What is a data paper
A data paper is a scholarly journal publication whose primary purpose is to describe a dataset or a group of datasets, rather than to report a research investigation (
- to provide a citable journal publication that brings scholarly credit to data creators,
- to describe the data in a structured human-readable form, and
- to bring the existence of the data to the attention of the scholarly community.
The description should include several important elements (usually called metadata, or “description of data”) that document, for example, how the dataset was collected, which taxa it covers, the spatial and temporal ranges and regional coverage of the data records, provenance information concerning who collected and who owns the data, details of which software (including version information) was used to create the data, or could be used to view the data, and so on.
Most Pensoft journals welcome submission and publication of data papers, that can be indexed and cited like any other research article, thus bringing registration of priority, a permanent publication record, recognition, and academic credit to the data creators. In other words, the data paper is a mechanism to acknowledge efforts in authoring ‘fit-for-use’ and enriched metadata describing a data resource. The general objective of data papers in biodiversity science is to describe all types of biodiversity data resources, including environmental data resources.
An important feature of data papers is that they should always be linked to the published datasets they describe, and that link (a URL, ideally resolving a DOI) should be published within the paper itself. Conversely, the metadata describing the dataset held within data archives should include the bibliographic details of the data paper once that is published, including a resolvable DOI. Ideally, the metadata should be identical in the two places — the data paper and the data archive — although this may be difficult to achieve with some archive metadata templates, so that there may be two versions of the metadata. This is why referring to the the data paper DOI is so important.
How to write and submit a data paper
In principle, any valuable dataset hosted in a trusted data repository can be described in a data paper and published following these Guidelines. Each data paper consists of a set of elements (sections), some of which are mandatory and some not. An example of such a list of elements needed to describe primary biodiversity data is available in the section data papers Describing Primary Biodiversity Data below.
Sample data papers which can be used as illustration of the concept can be downloaded from several Pensoft journals, for example, ZooKeys (examples), or Biodiversity Data Journal (examples).
All claims in a data paper should be substantiated by the associated data. If the methodology is standard, please explain in what respects your data are unique and merit a publication in the form of a data paper.
Alternatively, if the methodology used to acquire the data differs significantly from established approaches, please consider submitting your data to an open repository and associating them with a standard or data paper, in which these methodologies can be more fully explained.
At the time of submission of the data paper manuscript, the data described should be freely available online in a public repository under a suitable data license, so that they can be peer-reviewed, retrieved anonymously for re-use, resampling and redistribution by anyone for any purpose, subject to one condition at most — that of proper attribution using scholarly norms (see the Data Publishing Licenses and How to Cite Data sections, above). The repository, or at least one public mirror thereof, should not be under the control of the submitting authors. The relevant data package DOIs or accession numbers, as well as any special instructions for acquiring and re-publishing the data, should be included in the submitted data paper manuscript.
The procedures for data retrieval should be described, along with the mechanisms for updating and correcting information. This can be achieved by referencing an existing description if that is up to date, citable in its exact version, and publicly accessible on the web.
All methodological details necessary to replicate the original acquisition of the raw data have to be included in the data paper, along with a description of all data processing steps undertaken to transform the raw data into the form in which the data have been deposited in the repository and presented in the paper. Authors should discuss any relevant sources of error and how these have been addressed.
In addition to data papers describing new data resources, data papers describing legacy data are also welcome, as long as the current version of these is publicly accessible and can be cited. If possible, authors should outline possible re-use cases, taking into account that future uses of the data might involve researchers from different backgrounds. We encourage the provision of tools to facilitate visualization and re-use of the data.
For primary biodiversity (species-by-occurrence) data, authors are strongly encouraged to use the data publishing workflow of the GBIF Integrated Publishing Toolkit (IPT), described below. From IPT, data manuscripts can be generated in rich text format (RTF) directly from the metadata (Fig.
A more universal and innovative approach is conversion of the Ecological Metadata Language (EML) file available from IPT or other data platforms, such as DataONE or LTER, into data paper manuscripts in the ARPHA Writing Tool (Fig.
Data Papers Describing Primary Biodiversity Data
Primary biodiversity data as defined by GBIF are "Digital text or multimedia data records detailing facts about the instance of occurrence of an organism, i.e. on the what, where, when, how and by whom of the occurrence and the recording".
Currently, the majority of primary biodiversity data consists of species-by-occurrence data records available from published sources and/or natural history collections. Other types of primary biodiversity data that merit publication are observational data and multimedia resources in biodiversity.
Authoring metadata through the GBIF Integrated Publishing Toolkit (IPT)
The GBIF Integrated Publishing Toolkit (IPT) facilitates authoring of metadata based on the GBIF Metadata Profile (GMP) that was developed to standardise how biodiversity data resources are described for discovery through the GBIF network. For further information, see the GBIF Metadata Profile, Reference Guide and GBIF Metadata Profile, How-to guide.
The GMP conforms to the Ecological Metadata Language (EML) specification with some additional terms drawn from the Natural Collections Descriptions (NCD) set of terms for describing natural history collections and the ISO 19139: North American Profile of ISO 19115:2033 — Geographic Information — Metadata. The GMP elements, together with their descriptions, are listed below.
The structure of a Data Paper largely resembles that of a standard research paper. However, it must contain several specific elements. These elements are listed in Table
Structure of a data paper and its mapping from GBIF IPT Metadata Profile elements.
|
Section/Sub-Section headings of the data paper describing primary biodiversity data |
Mapping from GBIF IPT Metadata Profile elements, and formatting instructions |
|
<TITLE> |
Derived from the ‘title’ element. Format: a centred sentence without a full stop (.) at the end. |
|
<Authors> |
Derived from the ‘creator’, ‘metadataProvider’ and ‘AssociatedParty’ elements. From these elements, combinations of ‘first name’ and ‘last name’ are derived, separated by commas(,). Corresponding affiliations of the authors are denoted with numbers (1, 2, 3,...) superscripted at the end of each last name. If two or more authors share the same affiliation, it will be denoted by use of the same superscript number. Format: centred. |
|
<Affiliations> |
Derived from the ‘creator’, ‘metadataProvider’ and ‘AssociatedParty’ elements. From these elements, combinations of ‘Organisation Name’, ‘Address’, ‘Postal Code’, ‘City’, ‘Country’ constitute the affiliation. |
|
<Corresponding authors> |
Derived from the ‘creator’ and ‘metadataProvider’ elements. From these elements, ‘first name’, ‘last name’ and ‘email’ are derived. Email addresses are written in parentheses (). In a case of more than one corresponding author, these are separated by commas. If both creator and metadataProvider is the same, the creator is denoted as the corresponding author. Format: indented from both sides. |
|
<Received, Revised, Accepted, and Published dates> |
These will be inserted manually by the Publisher of the data paper, to indicate the dates of original manuscript submission, revised manuscript submission, acceptance of manuscript and publication of the manuscript as a data paper in the journal. |
|
<Citation> |
This will be inserted manually by the Publisher of the data paper. It will be a combination of Authors, Year of data paper publication (in parentheses), Title, Journal Name, Volume, Issue number (in parentheses), and DOI of the data paper, in both native and resolvable HTTP format. |
|
<Abstract> |
Derived from the ‘abstract’ element. Format: indented from both sides. |
|
<Keywords> |
Derived from ‘keyword’ element. Keywords are separated by commas (,). |
|
<Introduction> |
Free text. |
|
<Taxonomic Coverage> |
Derived from the Taxonomic Coverage elements. These elements are ‘general taxonomic coverage description’, ‘taxonomicRankName’, ‘taxonomicRankValue’ and ‘commonName’. ‘TaxanomicRankName’ and ‘taxonomicRankValues’. |
|
<Spatial Coverage> |
Derived from the Spatial Coverage elements. These elements are ‘general geographic description’, ‘westBoundingCoordinate’, ‘eastBoundingCoordinate’, ‘northBoundingCoordinate’, ‘southBoundingCoordinate’. |
|
<Temporal Coverage> |
Derived from the Temporal Coverage elements namely, ‘beginDate’ and ‘endDate’. |
|
<Project Description> |
Derived from project elements as described in the GBIF Metadata Profile. These elements are ‘title’ of the project, ‘personnel’ involved in the project, ‘funding sources’, ‘StudyAreaDescription/descriptor’, and ‘designDescription’. |
|
<Natural Collections Description> |
Derived from project NCD elements as described in the GBIF Metadata profile. These elements are ‘parentCollectionIdentifier’, ‘collectionName’, ‘collectionIdentifier’, formationPeriod’, ‘livingTimePeriod’, ‘specimenPreservationMethod’, and ‘curatorialUnit’. |
|
<Methods> |
Derivedfrom methods elements as described in the GBIF Metadata Profile. These elements are ‘methodStep/description’, ‘Sampling/StudyExtent/ description’, ‘sampling/samplingDescription’, and ‘qualityControl/ description’. |
|
<Dataset descriptions> |
Derived from physical and other elements as described in the GBIF Metadata Profile. These elements are ‘objectName’, ‘characterEncoding ’,‘formatName’, ‘formatVersion’, ‘distribution/online/URL’ ,‘pubDate’, ‘language’, and ‘intellectualRights’. |
|
<Additional Information> |
Derived from ‘additionalInfo’ element. |
|
<References> |
Derived from ‘citation’ element. This element assumes a reference to a research article or a web link, cited in the metadata description. |
The GBIF Integrated Publishing Toolkit (IPT) makes it easy to share different types of biodiversity-related information: primary taxon occurrence data (also known as primary biodiversity data), taxon checklists, sample-based data, and general metadata about data sources. An IPT instance, as well as the data and metadata registered through the IPT, is connected to the GBIF Registry, indexed for access via the GBIF network and portal, and made accessible for public use.
The IPT is a server-side software tool that allows users to author metadata, map databases or upload text files that conform to the Darwin Core standard, to install extensions and vocabularies to allow for richer content and, ultimately, to register datasets for publication and sharing through GBIF. IPT operators undertake the responsibility of running an Internet server which should be maintained, namely, that it should remain online and be addressable. Any set of metadata can be downloaded from any IPT (version 2.0.2+) into RTF format in the form of a data paper manuscript (Fig.
Therefore, data authors have the following options:
- Install and run an IPT instance, registering it with GBIF.
- Use an account on the Pensoft IPT Data Hosting Centre at http://ipt.pensoft.net; if you do not have an account yet, please ask the journal's Editorial Office to open one for you.
- Approach any other existing IPT operator and seek to host data through them.
GBIF provides a list of existing IPT installations supporting the authoring of data papers and a user manual for the IPT.
Once you have decided to publish your data and generate a data paper manuscript through the GBIF IPT, please consider the following simple rules:
- The metadata within one IPT generated archive must describe only one core set of biodiversity data (e.g., either occurrence data, a taxon checklist, or sample data), that is uploaded through the IPT, indexed in the GBIF Data Portal, and published in Darwin Core Archive Format. The IPT will generate an RTF manuscript that will describe the core dataset. The link to the core dataset will appear in your manuscript under the heading “Data published through GBIF”.
- Additional datasets that relate to the core one, e.g., ecological or environmental data, can also be briefly described within the same resource and linked through the "External links" field of the IPT. Those datasets will appear in the section “External datasets” of your manuscript.
- It is possible to open a resource and enter the respective metadata for it without upload of a core dataset. This option should be used to describe a dataset that has been already uploaded to a repository (e.g., data previously indexed through GBIF for which you have a GBIF link). In this case, you will need to insert the link(s) to the dataset(s) into the "External links" field of the IPT.
- The option explained in point 3 above can also be used to describe non-digital natural history collections.
- We strongly recommend uploading a core set of biodiversity data through the IPT Darwin Core Archive format, which facilitates not only publication of your data but also its easy sharing and integration with other data, hence its re-use and dissemination.
Generation of data paper manuscripts in RTF using the GBIF IPT
As described in the previous section, data creators will be able to author data paper manuscripts in various ways. However, to lower the technical barrier and make the process easy to adopt, a conversion tool to automatically export metadata to an RTF manuscript is available in IPT 2.0.2+. The step-by-step process for generating a data paper manuscript from the metadata is described below:
- The Data Creator completes the metadata for a biodiversity resource dataset using the metadata editor in IPT 2.0.2+. IPT assigns the Persistent Identifier to the authored metadata.
- Once the metadata are complete to the best of the author's ability, a data paper manuscript may be generated automatically from these metadata using the automated tool available within IPT 2.0.2+ (for RTF download from the dataset webpage, see Fig.
5 ). - The author checks the created manuscript, completing the textual Introduction or other appropriate sections, and then submits it for publication in the data paper section of an appropriate Pensoft journal through the online submission system (except for the Biodiversity Data Journal, One Ecosystem or RIO Journal, as these accept manuscripts in a different format).
- The manuscript undergoes peer review according to the journal's policies and the Guidelines for Reviewers of data paper (below). After review, and in case of acceptance, the manuscript is returned to the author by the editor along with the reviewers' and editorial comments for any required pre-publication modifications.
- The corresponding author inserts all accepted corrections or additions recommended by the reviewers and the editor in the metadata (not the manuscript of the paper), thereby improving the metadata for the data resource itself. Once the metadata have been improved, the final revised version of the data paper manuscript can then be created using the same automated metadata-to-manuscript conversion tool within IPT 2.0.2+ h was used to create the initally submitted draft (RTF download, see Fig.
5 ). - After manual re-insertion of the text of the Introduction, the revised data paper manuscript can then be submitted to the journal for final review and subsequent acceptance decision.
- Once the manuscript is accepted, it goes to a proofing stage, at which point submission, revision, acceptance and publication dates are added by the publisher, and a Digital Object Indentifier (DOI) is assigned to the data paper. This facilitates persistent accessibility of the online scholarly publication.
- Once the final proofs are approved by the author, the data paper is published in four different formats: (a) semantically enhanced HTML to provide interactive readings and links to external resources, (b) PDF, (c) final published XML to be archived in PubMedCentral and other archives to facilitate machine readability and future data mining, and eventually also (d) print format identical to the PDF version.
- After publication, the DOI of the data paper is linked with the Persistent Identifier of the metadata document registered in the GBIF Registry, which is given in the data paper. This provides multiple cross-linking between the data resource, its corresponding metadata and the corresponding data paper.
- Depending on the journal's policies and scope, the published data paper will be actively disseminated through the world's leading indexers and archives, including Web of Knowledge (ISI), PubMedCentral, Scopus, Zoological Record, Google Scholar, CAB Abstracts, Directory of Open Access Journal(DOAJ), EBSCOHost, and others.
Automated generation of data paper manuscripts from Ecological Metadata Language (EML) files
An innovative approach, similar to the that which converts EML metadata into RTF, is the direct conversion of an EML file (supported versions 2.2.0 and 2.2.1) downloaded from GBIF IPT (Fig.
- The users of ARPHA need to save a dataset's metadata as an EML file (versions 2.1.1 and 2.1.0, support for other versions is being continually updated) from the website of the respective data provider (see Fig.
5 as an example using the GBIF's Integrated Publishing Toolkit (GBIF IPT)). Some leading data portals that provide such EML files are GBIF (EML download possible both from IPT and from the portal see Fig.5 ), DataONE, and the LTER Network. - Click on the "Start a manuscript" button in AWT and then select "Biodiversity Data Journal" and the "Data paper (Biosciences)" template (Fig.
7 ). - Upload this file via the "Import a manuscript" function on the AWT interface (Fig.
8 ). - Continue with updating and editing and finally submit your manuscript inside AWT.
Data Papers Describing Ecological and Environmental Data
Metadata descriptions of primary biodiversity data used in the GBIF Metadata Proifile (GMP) and the Integrated Publishing Toolkit (IPT) are based primarily on the Ecological Metadata Language (EML) Specification. Therefore, the same basic elements and the overall data paper structure explained in the previous section can also be used to describe ecological and environmental data. As a result, data papers for ecological and environmental data will have a basic structure similar to that of papers on primary biodiversity data. Authors are encouraged to include additional elements (sections) in the manuscripts if they expect this to improve the description of the specifics of their environmental and ecological data. The main difference is that ecological and environmental data cannot be processed through the GBIF IPT and hence they should be deposited in another public data hosting centre listed in the section Open Data Repositories, for example DataONE, LTER Network, PANGAEA or Dryad.
Authors intending to publish data papers describing ecological and environmental data are advised to use the following steps:
- Deposit your data in an ISO-certified public (international or institutional) repository.
- Write a data paper manuscript following the structure of the sample data paper, adding additional elements/sections to the manuscript if these are necessary to describe the specifics of your dataset(s).
- Add the permanent link(s) in the manuscript to the particular dataset(s) hosted in the repository you have chosen.
- Submit the data paper to an appropriate Pensoft journal.
- Once the paper is accepted and published, enter the bibliographic reference and the DOI of the data paper in the relevant metadata field of your data package in the repository that hosts your data.
Alternatively, EML metadata files (versions 2.2.0 and 2.2.1) hosted in DataONE and LTER can automatically be converted into а data paper manuscript using the ARPHA Writing Tool import workflow described in the previous section (see also
Data Papers Describing Genomic Data
Pensoft journals require, as a condition for publication, that genome data supporting the results in the paper should be archived in an appropriate public archive, and accession numbers must be included in the final version of the paper. Sufficient additional metadata (such as sample locations, individual identities, etc.) should also be provided to allow easy repetition of analyses presented in the paper. For best practice in following community metadata standards, see the many data-type specific standards and checklists provided by the Genomic Standards Consortium (particularly the MIxS standards, as described by
DNA sequence data should be archived in GenBank or another public database of the INSDC consortium. Expression data should be submitted to the Gene Expression Omnibus or an equivalent database, whereas phylogenetic trees should be submitted to TreeBASE. More idiosyncratic data, such as microsatellite allele frequency data, can be archived in a more flexible digital data repository such as Dryad or Knowledge Network for Biocomplexity (KNB).
Barcode Data Release Papers
Barcode-of-Life COI (mitochondrial encoded cytochrome oxidase 1) genome data can be published in a form of a Data Paper, as has been announced by the Consortium for the Barcode of Life (CBOL) and illustrated by some published sample papers (
Definition: A BARCODE Data Release Paper is a short manuscript that announces and documents the public release to a member of the International Nucleotide Sequence Data Collaboration (INSDC, which includes GenBank, ENA,and DDBJ) of a significant body of data records that meet the BARCODE data standards.
Contents: BARCODE Data Release Papers are meant to announce and document the public availability of a significant body of new DNA barcodes. The barcode records should therefore be a coherent set of records that provides noteworthy new research capabilities for a taxonomic group, ecological assemblage or specified geographic region. Authors should explain the rationale for creating a comprehensive library of BARCODE data for that taxonomic group, ecological habitat, and/or geographic region. If the data have been collected as part of a larger, longer-term research project, the manuscript should explain the wider project and its planned use of the data for taxonomic, biogeographic, evolutionary, and/or applied research, or for other purposes.
The BARCODE Data Release Paper manuscript should describe:
- The scope of taxonomic, ecological, and geographic coverage;
- The sources of voucher specimens;
- The sampling and laboratory protocols used;
- The processes used to identify the species to which voucher specimens belong.
The manuscript should provide summaries of data density and quality such as those shown in Table
Suggested data fields for a BARCODE Data Release Paper
|
Average number of records per species |
|
| Range of records per species | Min-Max |
| Average sequence length (and Min/Max) | |
| Range of intraspecific variation* | Min-Max |
| Median variation within species* | X% |
| Range of divergence between closest species-pairs** | Min-Max |
| Median divergence between closest species-pairs** | |
* Calculated as the arithmetic average of all K2P distances between specimens in each species.
** Closest species pairs refers to each species and the species with which it has the least divergent barcode sequence. The true phylogenetic sister-species may not be included in the dataset, and could have a lower interspecies divergence.
Manuscripts should also include an Appendix with a table that presents:
- The taxonomic identification (a formal species name or a provisional species label in a public database);
- The collecting locality to a reasonable level of precision;
- The voucher specimen identifier in the format required in the BARCODE data standard;
- The accession number in GenBank, EMBL or DDBJ; and
- The Barcode of Life Data Systems (BOLD) record number (optional).
Review Criteria: In addition to the general Guidelines for Reviewers listed in the next section, CBOL recommends that reviewers use the following evaluation criteria for BARCODE Data Release Papers, and suggests that authors anticipate such evaluation:
- Data quality: All data records should meet the BARCODE data standards agreed to by CBOL and INSDC. The manuscript should demonstrate the effectiveness of the BARCODE records in distinguishing species, as well as pointing out limitations of the BARCODE data for species identification.
- Significance of the data records: The data records being released should represent a significant addition to the public knowledge base. The manuscript should demonstrate the significance of the new records relative to the previously released BARCODE data for that combination of taxonomic group, ecological habitat, and geographic region. Manuscripts that announce the release of the first BARCODE records representing a higher proportion of species in a taxonomic group will have higher significance.
- Relevance to other research programs and societal applications: BARCODE data release manuscripts will be considered more relevant if they treat taxonomic groups, ecological habitats, and geographic regions that are connected with other basic research programs in evolutionary biology or ecology, or are components of applied research for socioeconomic reasons (e.g., agriculture, food safety, conservation, environmental monitoring, public health).
- Documentation and accessibility: The voucher specimens and their associated data and metadata will be valuable resources for the research community. The data table in the Appendix must provide links to the voucher specimens and taxonomic identification, as well as the INSDC Accession Numbers. Reviewers will evaluate the degree to which voucher specimens are available in permanent repositories (as opposed to private research collections) and degree to which taxonomic identifications are documented in published or other resources. Provisional non-Linnean taxonomic labels may be used, but they should be linked to online databases that document the author's concept of the taxonomic unit.
Software Description Papers
An increasing number of software tools also merit description in scholarly publications. The structure of the data paper proposed below for such software tools is largely based on the Description of a Project (DOAP) RDF schema and XML vocabulary developed by Edd Dumbill to describe software projects, in particular those that are open-source. The main difference, however, is that the data paper aims at the description of the software product and not of the software source code; data papers of this kind are addressed mainly to end users of the software and less to developers and software engeneers.
Software citation principles have been developed by the FORCE11 Software Citation Working Group based on an adaptation of the FORCE11 Data Citation Principles. The six principles are abstracted here (
- Importance: Software should be considered a legitimate and citable product of research.
- Credit and Attribution: Software citations should facilitate giving scholarly credit and normative and legal attribution to all contributors to the software.
- Unique Identification: A software citation should include a method for identification that is machine actionable, globally unique, interoperable, and recognizable.
- Persistence: Unique identifiers and metadata describing the software and its disposition should persist.
- Accessibility: Software citations should facilitate access to the software itself and to its associated metadata, documentation, data, and other materials.
- Specificity: Software citations should facilitate identification of, and access to, the specific version of software that was used.
Based on an analysis of several use cases such as publishing a software paper or publishing papers that cite software, basic metadata requirements were identified: unique identifier, software name, author(s), contributor role, version number, release date, location/repository, indexed citations, software license, description, keywords.
While the provision of detailed specifications and recommendations around metadata standards were beyond the scope of the working group, DOAP is mentioned together with some other more recent community initiatives. It is expected that a new working group will take these software citation principles forward by supporting potential implementers and developing metadata standards, following the example of the FORCE11 Data Citation Working Group (
According to DOAP, major properties of a software tool description include elements such as homepage, developer, programming language and operational system. Other properties include: Implements specification, anonymous root, platform, browse, mailing list, category, description, helper, tester, short description, audience, screenshots, translator, module, documenter, wiki, repository, name, repositorylocation, language, service endpoint, created, download mirror, vendor, old homepage, revision, download page, license, bug database, maintainer, blog, file-release, and release.
A basic version of a DOAP description can be generated using an online tool called doapamatic.
A sample structure of a Software Description paper was introduced and used by Pensoft since 2011 (
Metadata elements (based on EML and DOAP) to be included in a data paper describing a software tool
|
Section/Sub-Section headings of the Software Description paper |
Mapping from the available EML and DOAP metadata elements; a few other elements have been added to provide a better mapping to the data paper structure, with formatting guidelines |
|
<TITLE> |
Derived from the ‘name’ element. This must be extended to a concise description of the software tool and its implementation, e.g.: “BioDiv, a web-based tool for calculation of biodiversity indexes”. Format: This is a centred sentence without full stop (.) at the end. |
|
<Authors> |
Derived from the ‘developer’, ‘maintainer’ and eventually ‘helper’, ‘tester’, and ‘documenter’. From these elements, combinations of ‘first name’ and ‘last name’ are derived, separated by commas (,). Corresponding affiliations of the authors are denoted with numbers (1, 2, 3,...) in superscript at the end of each last name. If two or more authors share same affiliation, it will be denoted by use of the same superscript number. Format: centred. |
|
<Affiliations> |
Derived from the ‘developer’, ‘maintainer’ and ‘helper’. From these elements, combinations of ‘Organisation Name’, ‘Address’, ‘Postal Code’, ‘City’, and ‘Country’ will constitute the affiliation. |
|
<Corresponding authors> |
Derived from any of the ‘developer’, ‘maintainer’, ‘helper’, ‘tester’, and ‘documenter’ elements. From these elements ‘first name’, ‘last name’ and ‘email’ are derived. Email addresses are written in parentheses (). In case of more than one corresponding author, these are separated by commas. Format: indented from both sides. |
|
<Received, Revised, Accepted, and Published dates> |
These will be inserted manually by the Publisher of the data paper to indicate the dates of original manuscript submission, revised manuscript submission, acceptance of manuscript and publishing of the manuscript as data paper in the journal. |
|
<Citation> |
This will be inserted manually by the Publisher of the data paper. It will be a combination of Authors, Year of data paper publication (in parentheses), Title, Journal Name, Volume, Issue number (in parentheses), and DOI of the data paper in both native and resolvable HTTP format. |
|
<Abstract> |
Derived from the ‘short description’ element. Format: indented from both sides. |
|
<Keywords> |
Keywords should reflect most important features of the tool and areas of implementation, and should be separated by commas (,). |
|
<Introduction> |
Free text. |
|
<Project Description> |
Derived from ‘description’ element; if applicable, it should also include sub-elements such as ‘title’ of the project, ‘personnel’ involved in the project, ‘funding’ sources’, and other appropriate information. |
|
<Web Location (URIs)> |
Derived from the elements ‘homepage’, ‘wiki’, ‘download page’, ‘download mirror’, ‘bug database’, ‘mailing list’, ‘blog’, ‘vendor’ |
|
<Technical specification> |
Derived from the elements ‘platform’, ‘programming language’, ‘operational system’ (if OS-specific), ‘language’ , ‘service endpoint’ |
|
<Repository> |
Derived from the elements ‘repository type’ (CVS, SVN, Arch, BK), ‘repository browse uri’ (CVS, SVN, BK), ‘repository location’) SVN, BK, Arch), ‘repository module’ (CVS, Arch), ‘repository anonymous root’ (CVS) |
|
<License> |
Derived from the ‘license’ element |
|
<Implementation> |
Derived from ‘Implements specification’ and ‘audience’ elements; please remember that this section is of primary interest to end users, and should be written in detail, if possible including use cases, citations and links. |
|
<Additional Information> |
Any kind of helpful additional information may be included. |
|
<Acknowledgement> |
Lists all acknowledgments at the authors' discretion. |
|
<References> |
Includes literature references and web links cited in the text. |
Guidelines for Reviewers and Editors
Data papers describing data resources — or manuscripts linked to open data resources that underpin the scientific analyses — that are submitted to Pensoft journals will be subjected to peer review according to the respective journal's policies (e.g., conventional pre-publication anonymous, non-anonymous, or entirely open and public, including post-publication review) as a routine method to enhance the completeness, truthfulness and accuracy of the descriptions of the relevant data resources, thereby improving their use and uptake. A specific feature of the ARPHA-XML journal publishing workflow used by the Biodiversity Data Journal, Research Ideas and Outcomes (RIO Journal), One Ecosystem, and others, is the so called pre-submission peer-review which can be organized by the authors or the journal's editorial office still during the authoring process in the ARPHA Writing Tool.
Peer review of data papers is expected to evaluate the completeness and quality of the dataset(s) description (metadata), as well as the publication value of data. This may include the appropriateness and validity of the methods used, compliance with applicable standards during collection, management and curation of data, and compliance with appropriate metadata standards in the description of the data resources. In order to allow for accuracy and usefulness, metadata needs to be as complete and descriptive as possible.
Reviewers will consider the following aspects of (a) the quality of the manuscript, (b) the quality of the data, and (c) the consistency between the description within the data paper and the repository-held metadata relating the data resource itself.
Peer review of the data is rather problematic in the current scholarly publishing practice. There are several reasons for that:
- Authors are not sufficiently trained in and accustomed to the good practices of formatting and describing their data.
- Reviewers do not pay sufficient attention to data reviews. A proper review of large datasets may appear merely impossible due to the volume of work.
- Editors are not sufficiently experienced in data review, which often requires specific training in data management.
- Data are of different types and specificities, which imposes additional problems to find suitable reviewers or editors.
- Data standards to consider as a "rule-to-follow" are at different levels of development and adoption by different communities.
Several Pensoft journals offer an additional service for auditing and correcting data, which might be a solution for those authors or their institutions who really care about data quality and re-use.
Best practice recommendations for evaluating data papers or manuscripts that are submitted together with the underlying data are summarised below.
Quality of the Manuscript
- Does the manuscript conform to the focus and scope of this journal?
- Does the manuscript contain unpublishable — for example fraudulent or pseudoscientific — content?
- Does the manuscript contain sufficiently detailed information to merit publication?
- Do the title, abstract and keywords accurately reflect the contents of the manuscript?
- Is the manuscript internally consistent and suitably organized?
- Is the manuscript written in grammatically and stylistically correct English?
- Are the methods relevant to the study and adequately described?
- Did the authors cite most of the literature pertinent to the subject?
- Are relevant non-textual media (e.g. tables, figures, audio, video) used to an appropriate extent and in a suitable manner?
- Have abbreviations and symbols been properly defined?
- Are the illustrations of sufficient quality?
- Does the manuscript put the data resource being described properly into the context of prior research, citing pertinent articles and datasets?
- Are conflicts of interest, relevant permissions and other ethical issues addressed in an appropriate manner?
Quality of the Data
- Are the data freely and openly available under an appropriate Creative Commons license or waiver?
- Is the repository to which the data are submitted appropriate for the nature of the data?
- Are the data completely and consistently recorded within the dataset(s)?
- Does the data resource cover scientifically important and sufficiently large region(s), time period(s) and/or group(s) of taxa to be worthy of a separate publication?
- Are the data consistent internally and described using applicable standards (e.g. in terms of file formats, file names, file size, units and metadata)?
- Are the methods used to process and analyse the raw data, thereby creating processed data or analytical results, sufficiently well documented that they could be repeated by third parties?
- Are the data plausible, given the protocols? Authors are encouraged to report any tests undertaken to address this point.
Consistency between Manuscript and Data
- Does the manuscript provide an accurate description of the data?
- Does the manuscript properly describe how to access the data?
- Are the methods used to generate the data (including calibration, code and suitable controls) described in sufficient detail?
- Is the dataset sufficiently unique to merit publication as a data paper?
- Are the use cases described in the data paper consistent with the data presented? Would other possible use cases merit comment in the paper?
- Have possible sources of error been appropriately addressed in the protocols and/ or the paper?
- Is anything missing in the manuscript or the data resource itself that would prevent replication of the measurements, or reproduction of the figures or other representations of the data?
- Are all claims made in the manuscript substantiated by the underlying data?
Acknowledgements
We thank several colleagues who commented or contributed to an earlier version of the draft: Tim Robertson and Kyle Braak (GBIF), Todd Vision and Peggy Schaeffer (Dryad). We also thank all our authors, reviewers, editors and partners for their support in testing and using these data publishing guidelines and workflows. Special thanks are due to Donat Agosty, Terry Catapano, Guido Sautter and Willi Egloff from Plazi (Switzerland) for the years-long successful collaboration, and friendship, and to Robert Mesibov (Tasmania) and Florian Wetzel (Museum für Naturkunde, Berlin) who provided open pre-submission peer reviews to the manuscript.
Funding program
The present guidelines were elaborated through the FP7 funded project EU BON: Building the European Biodiversity Observation Network, grant agreement ENV30845, and constitute Milestone MS842. V. Senderov's PhD is financed through the EU Marie-Sklodovska-Curie Program BIG4 project, Grant Agreement Nr. 642241.
References
-
Taxonomic information exchange and copyright: the Plazi approach.BMC Research Notes2:53. https://doi.org/10.1186/1756-0500-2-53
-
A Proposed Standard for the Scholarly Citation of Quantitative Data.D-Lib Magazine13(3):1‑11. URL: https://ssrn.com/abstract=1081955
-
An introduction to the joint principles for data citation.Bulletin of the American Society for Information Science and Technology41(3):43‑45. https://doi.org/10.1002/bult.2015.1720410313
-
Linking multiple biodiversity informatics platforms with Darwin Core Archives.Biodiversity Data Journal2:e1039. https://doi.org/10.3897/bdj.2.e1039
-
How to Cite Datasets and Link to Publications. DCC How-to Guides. Edinburgh: Digital Curation Centre.http://www.dcc.ac.uk/resources/how-guides. Accessed on: 2017-9-02.
-
BioMed Central's Position Statement on Open Data. https://blogs.biomedcentral.com/wp-content/image_archive/opendatastatementdraft.pdf
-
Data journals: A survey.Journal of the Association for Information Science and Technology66(9):1747‑1762. https://doi.org/10.1002/asi.23358
-
Species Conservation Profiles compliant with the IUCN Red List of Threatened Species.Biodiversity Data Journal4:e10356. https://doi.org/10.3897/bdj.4.e10356
-
TaxPub: An Extension of the NLM/NCBI Journal Publishing DTD for Taxonomic Descriptions.Journal Article Tag Suite Conference (JATS-Con) Proceedings 2010.National Center for Biotechnology Information (US),BethesdaURL: https://www.ncbi.nlm.nih.gov/books/NBK47081/
-
Models for financial sustainability of biological databases and resources.Database2009:bap017‑bap017. https://doi.org/10.1093/database/bap017
-
The data paper: a mechanism to incentivize data publishing in biodiversity science.BMC Bioinformatics12:S2. https://doi.org/10.1186/1471-2105-12-s15-s2
-
Towards a data publishing framework for primary biodiversity data: challenges and potentials for the biodiversity informatics community.BMC Bioinformatics10:S2. https://doi.org/10.1186/1471-2105-10-s14-s2
-
The International Nucleotide Sequence Database Collaboration.Nucleic Acids Research44:D48‑D50. https://doi.org/10.1093/nar/gkv1323
-
Out of Cite, Out of Mind: The Current State of Practice, Policy, and Technology for the Citation of Data.Data Science Journal12:CIDCR1‑CIDCR75. https://doi.org/10.2481/dsj.osom13-043
-
Aichi Biodiversity Target. https://www.cbd.int/sp/targets/. Accessed on: 2017-2-22.
-
Motivating Online Publication of Data.BioScience59(5):418‑427. https://doi.org/10.1525/bio.2009.59.5.9
-
Biodiversity data should be published, cited, and peer reviewed.Trends in Ecology & Evolution28(8):454‑461. https://doi.org/10.1016/j.tree.2013.05.002
-
A Data Citation Roadmap for Scientific Publishers.bioRxivhttps://doi.org/10.1101/100784
-
Best Practices for Data Sharing in Phylogenetic Research.PLoS Currentshttps://doi.org/10.1371/currents.tol.bf01eff4a6b60ca4825c69293dc59645
-
Adventures in data citation: sorghum genome data exemplifies the new gold standard.BMC Research Notes5(1):223. https://doi.org/10.1186/1756-0500-5-223
-
Biodiversity research in the “big data” era: GigaScience and Pensoft work together to publish the most data-rich species description.GigaScience2(1): . https://doi.org/10.1186/2047-217x-2-14
-
Experiences in integrated data and research object publishing using GigaDB.International Journal on Digital Librarieshttps://doi.org/10.1007/s00799-016-0174-6
-
EU BON Policy Brief On Open Data.Zenodohttps://doi.org/10.5281/ZENODO.188391
-
Open exchange of scientific knowledge and European copyright: The case of biodiversity information.ZooKeys414:109‑135. https://doi.org/10.3897/zookeys.414.7717
-
Copyright and the Use of Images as Biodiversity Data.bioRxivhttps://doi.org/10.1101/087015
-
Data Policy Recommendations for Biodiversity Data. EU BON Project Report.Research Ideas and Outcomes2:e8458. https://doi.org/10.3897/rio.2.e8458
-
Geographic distibution of the rock-dwelling door-snail genus Montenegrina Boettger, 1877 (Mollusca, Gastropoda, Clausiliidae).V1.5.ZooKeys. Dataset/Ocurrences deposited in GBIF. URL: http://ipt.pensoft.net/resource?r=montenegrina&v=1.5
-
DNA Barcodes for Nearctic Auchenorrhyncha (Insecta: Hemiptera).PLoS ONE9(7):e101385. https://doi.org/10.1371/journal.pone.0101385
-
We Need Publishing Standards for Datasets and Data Tables.OECD Publishing White Paper1:1. https://doi.org/10.1787/603233448430
-
Creative Commons licenses and the non-commercial condition: Implications for the re-use of biodiversity information.ZooKeys150:127‑149. https://doi.org/10.3897/zookeys.150.2189
-
A decadal view of biodiversity informatics: challenges and priorities.BMC Ecology13(1):16. https://doi.org/10.1186/1472-6785-13-16
-
A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names.MycoKeys1:7‑20. https://doi.org/10.3897/mycokeys.1.2062
-
The need for an integrated biodiversity policy support process – Building the European contribution to a global Biodiversity Observation Network (EU BON).Nature Conservation6:49‑65. https://doi.org/10.3897/natureconservation.6.6498
-
Amendment of Articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication.ZooKeys219:1‑10. https://doi.org/10.3897/zookeys.219.3944
-
Criteria for the Trustworthiness of Data Centres.D-Lib Magazine17(1):0. https://doi.org/10.1045/january2011-klump
-
Changes to publication requirements made at the XVIII International Botanical Congress in Melbourne - what does e-publication mean for you?PhytoKeys6:5. https://doi.org/10.3897/phytokeys.6.1960
-
Innovative electronic publication in plant systematics: PhytoKeys and the changes to the “Botanical Code” accepted at the XVIII International Botanical Congress in Melbourne.PhytoKeys6:1. https://doi.org/10.3897/phytokeys.6.2063
-
A geographical distribution database of the genus Dysdera in the Canary Islands (Araneae, Dysderidae).ZooKeys625:11‑23. https://doi.org/10.3897/zookeys.625.9847
-
Data from: A geographical distribution database of the genus Dysdera in the Canary Islands (Araneae, Dysderidae).Dryad Digital Repositoryhttps://doi.org/10.5061/DRYAD.T63MN
-
Data Citation Synthesis Group: Joint Declaration of Data Citation Principles. San Diego CA: FORCE11. https://www.force11.org/group/joint-declaration-data-citation-principles-final
-
BioSharing: curated and crowd-sourced metadata standards, databases and data policies in the life sciences.Database : the journal of biological databases and curation2016https://doi.org/10.1093/database/baw075
-
Editorial.The International Journal of Robotics Research28(5):587‑587. https://doi.org/10.1177/0278364909104283
-
Implementation of TaxPub, an NLM DTD extension for domain-specific markup in taxonomy, from the experience of a biodiversity publisher.Journal Article Tag Suite Conference (JATS-Con) Proceedings.Bethesda (MD).National Center for Biotechnology Information (US)[Inen]. URL: https://www.ncbi.nlm.nih.gov/books/NBK100351/
-
Publication and dissemination of datasets in taxonomy: ZooKeys working example.ZooKeys11:1‑8. https://doi.org/10.3897/zookeys.11.210
-
Pensoft Data Publishing Policies and Guidelines for Biodiversity Data.Zenodo1:1‑34. https://doi.org/10.5281/zenodo.56660
-
Data publication and dissemination of interactive keys under the open access model.ZooKeys21:1‑17. https://doi.org/10.3897/zookeys.21.274
-
A common registration-to-publication automated pipeline for nomenclatural acts for higher plants (International Plant Names Index, IPNI), fungi (Index Fungorum, MycoBank) and animals (ZooBank).ZooKeys550:233‑246. https://doi.org/10.3897/zookeys.550.9551
-
ZooKeys, unlocking Earth’s incredible biodiversity and building a sustainable bridge into the public domain: From “print-based” to “web-based” taxonomy, systematics, and natural history. ZooKeys Editorial Opening Paper.ZooKeys1:1‑7. https://doi.org/10.3897/zookeys.1.11
-
DNA Barcodes for the Northern European Tachinid Flies (Diptera: Tachinidae).PLOS ONE11(11):e0164933. https://doi.org/10.1371/journal.pone.0164933
-
BARCODING: BOLD: The Barcode of Life Data System (http://www.barcodinglife.org).Molecular Ecology Notes7(3):355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
-
A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System.PLoS ONE8(7):e66213. https://doi.org/10.1371/journal.pone.0066213
-
Data Citation of Evolving Data: Recommendations of the RDA Working Group on Data Citation (WGDC).Research Data Alliancehttps://doi.org/10.15497/RDA00016
-
A DNA barcode library for ground beetles (Insecta, Coleoptera, Carabidae) of Germany: The genus Bembidion Latreille, 1802 and allied taxa.ZooKeys592:121‑141. https://doi.org/10.3897/zookeys.592.8316
-
About RDA. https://www.rd-alliance.org/about-rda. Accessed on: 2017-9-02.
-
The GBIF Integrated Publishing Toolkit: Facilitating the Efficient Publishing of Biodiversity Data on the Internet.PLoS ONE9(8):e102623. https://doi.org/10.1371/journal.pone.0102623
-
PASSIFOR: A reference library of DNA barcodes for French saproxylic beetles (Insecta, Coleoptera).Biodiversity Data Journal3:e4078. https://doi.org/10.3897/bdj.3.e4078
-
BioSharing Overview.Nature Precedingshttps://doi.org/10.1038/npre.2011.5936
-
The Global Registry of Biodiversity Repositories: A Call for Community Curation.Biodiversity Data Journal4:e10293. https://doi.org/10.3897/bdj.4.e10293
-
Project description: DNA barcodes of bird species in the national museum of natural history, smithsonian institution, USA.ZooKeys152:87‑92. https://doi.org/10.3897/zookeys.152.2473
-
Online direct import of specimen records into manuscripts and automatic creation of data papers from biological databases.Research Ideas and Outcomes2:e10617. https://doi.org/10.3897/rio.2.e10617
-
Software Citation Principles.PeerJ Preprintshttps://doi.org/10.7287/PEERJ.PREPRINTS.2169V4
-
Beyond dead trees: integrating the scientific process in the Biodiversity Data Journal.Biodiversity Data Journal1:e995. https://doi.org/10.3897/bdj.1.e995
-
Data publication: towards a database of everything.BMC Research Notes2(1):113. https://doi.org/10.1186/1756-0500-2-113
-
Achieving human and machine accessibility of cited data in scholarly publications.PeerJ Computer Science1:e1. https://doi.org/10.7717/peerj-cs.1
-
Eupolybothrus cavernicolus Komerički & Stoev sp. n. (Chilopoda: Lithobiomorpha: Lithobiidae): the first eukaryotic species description combining transcriptomic, DNA barcoding and micro-CT imaging data.Biodiversity Data Journal1:e1013. https://doi.org/10.3897/bdj.1.e1013
-
Sharing and re-use of phylogenetic trees (and associated data) to facilitate synthesis.BMC Research Notes5(1):574. https://doi.org/10.1186/1756-0500-5-574
-
Revision of the Malagasy genus Trichoteleia Kieffer (Hymenoptera, Platygastroidea, Platygastridae).ZooKeys80:1‑126. https://doi.org/10.3897/zookeys.80.907
-
Data issues in the life sciences.ZooKeys150:15‑51. https://doi.org/10.3897/zookeys.150.1766
-
Toward Implementation of the Global Earth Observation System of Systems Data Sharing Principles.Data Science Journal8:1‑91. URL: https://www.jstage.jst.go.jp/article/dsj/8/0/8_35JSL201/_pdf
-
A natural language programming solution for executable papers.Procedia Computer Science4:678‑687. https://doi.org/10.1016/j.procs.2011.04.071
-
Catalogue of the type specimens of Polycestinae (Coleoptera: Buprestidae) from research collections of the Zoological Institute, Russian Academy of Sciences.Zoological Institute, Russian Academy of Sciences, St. Petersburg, deposited in GBIFhttps://doi.org/10.15468/C3EORK
-
The roles and contributions of Biodiversity Observation Networks (BONs) in better tracking progress to 2020 biodiversity targets: a European case study.Biodiversity16:137‑149. https://doi.org/10.1080/14888386.2015.1075902
-
Darwin Core: An Evolving Community-Developed Biodiversity Data Standard.PLoS ONE7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
-
The FAIR Guiding Principles for scientific data management and stewardship.Scientific Data3:160018. https://doi.org/10.1038/sdata.2016.18
-
Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications.Nature Biotechnology29(5):415‑420. https://doi.org/10.1038/nbt.1823