|
Research Ideas and Outcomes :
NSF Grant Proposal
|
|
Corresponding author:
Received: 22 Sep 2016 | Published: 30 Sep 2016
© 2016 Nico Franz, Edward Gilbert, Bertram Ludäscher, Alan Weakley
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Franz N, Gilbert E, Ludäscher B, Weakley A (2016) Controlling the taxonomic variable: Taxonomic concept resolution for a southeastern United States herbarium portal. Research Ideas and Outcomes 2: e10610. https://doi.org/10.3897/rio.2.e10610
|
|
Executive summary
Overview. Taxonomic names are imperfect identifiers of specific and sometimes conflicting taxonomic perspectives in aggregated biodiversity data environments. The inherent ambiguities of names can be mitigated using syntactic and semantic conventions developed under the taxonomic concept approach. These include: (1) representation of taxonomic concept labels (TCLs: name sec. source) to precisely identify name usages and meanings, (2) use of parent/child relationships to assemble separate taxonomic perspectives, and (3) expert provision of Region Connection Calculus articulations (RCC–5: congruence, [inverse] inclusion, overlap, exclusion) that specify how data identified to different-sourced TCLs can be integrated. Application of these conventions greatly increases trust in biodiversity data networks, most of which promote unitary taxonomic 'syntheses' that obscure the actual diversity of expert-held views. Better design solutions allow users to control the taxonomic variable and thereby assess the robustness of their biological inferences under different perspectives. A unique constellation of prior efforts – including the powerful Symbiota collections software platform, the Euler/X multi-taxonomy alignment toolkit, and the "Weakley Flora" which entails 7,000 concepts and more than 75,000 RCC–5 articulations – provides the opportunity to build a first full-scale concept resolution service for SERNEC, the SouthEast Regional Network of Expertise and Collections, currently with 60 member herbaria and 2 million occurrence records.
Intellectual merit. We have developed a multi-dimensional, step-wise plan to transition SERNEC's data culture from name- to concept-based practices. (1) We will engage SERNEC experts through annual, regional workshops and follow-up interactions that will foster buy-in and ultimately the completion of 12 community-identified use cases. (2). We will leverage RCC–5 data from the Weakley Flora and further development of the Euler/X logic reasoning toolkit to provide comprehensive genus- to variety-level concept alignments for at least 10 major flora treatments with highest relevance to SERNEC. The visualizations and estimated > 1 billion inferred concept-to-concept relations will effectively drive specimen data integration in the transformed portal. (3) We will expand Symbiota's taxonomy and occurrence schemas and related user interfaces to support the new concept data, including novel batch and map-based specimen determination modules, with easy output options in Darwin Core Archive format. (4) Through combinations of the new technology, enlisted taxonomic expertise, and SERNEC's large image resources, we will upgrade minimally 80% of all SERNEC specimen identifications from names to the narrowest suitable TCLs, or add "uncertainty" flags to specimens needing further study. (5) We will utilize the novel tools and data to demonstrate how controlling for the taxonomic variable in 12 use cases variously drives the outcomes of evolutionary, ecological, and conservation-based research hypotheses.
Broader impacts. Our project is focused on just one herbarium network, but the potential impact is as wide as Darwin Core or even comparative biology. We believe that trust in networked biodiversity data depends on open and dynamic system designs, allowing expert access and resolution of multiple conflicting views that reflect the complex realities of ongoing taxonomic research. Taking well over 1 million SERNEC records from name- to TCL-resolution will show that "big" specimen data can pass the credibility threshold needed to validate the substantive data mobilization investment. We will mentor one postdoctoral researcher (UNC), two Ph.D. students (ASU, UIUC), and at least 15 undergraduate students (ASU). Each of our workshops will capacitate 10-15 SERNEC experts, who in turn can recruit colleagues and students at their home collections. We will incorporate the project theme and use cases into undergraduate courses taught at six institutions and reaching an estimated 300-500 students annually (10-40% minority students). At each institution, project members will make a systematic effort to recruit new students from underrepresented groups. Our group's leadership of Symbiota (with close ties to iDigBio), SERNEC, and local biodiversity projects and centers will further promote the new data culture. We will create a feature story "Where do plant species occur?" for ASU's popular "Ask A Biologist" website, and a series of undergraduate student-led "How-To" videos that illustrate the use case workflows, including the creation of multi-taxonomy alignments.
Keywords
Aggregation, concept taxonomy, conflict, flora, herbarium, logic, reasoning, Region Connection Calculus, specimens, synthesis
List of participants
Mac Alford, Mark Fishbein, Alan Franck, Nico Franz, Edward Gilbert, Michael Lee, Zack Murrell, Bertram Ludäscher, Pamela Soltis, Alan Weakley.
Data management plan
Types of Data Produced
Data to be produced and managed for the project include: (1a) Software code written for the Symbiota content management system (primarily written in PHP and with heavy use of JavaScript libraries; and connecting to the open source MariaDB SQL database platform) and (1b) for the Euler/X logic reasoning toolkit (primarily written in Python); (2) specimen occurrence records (with new identifications) managed in the Symbiota-operated SERNEC herbarium portal, and formatted in compliance (where possible; see details below) with the Taxonomic Working Group (TDWG) -endorsed Darwin Core (DwC) and Taxonomic Concept Transfer Schema (TCS) standards (https://github.com/tdwg); and (3) Euler/X toolkit input/output files, presently stored in simple .csv, .gv (GraphViz), .pdf, .txt, and .yaml file formats. We will also (4) author web posts (.html) and instructional videos (.mp4) (see Broader Impacts).
Data and Metadata Standards
The Symbiota-based SERNEC portal occurrence data are fully Darwin Core-compatible. These data can be bundled through easy-to-use platform functions to yield Darwin Core Archive files for wider sharing. We note, however, that Darwin Core does not presently support all syntactic and semantic conventions of the taxonomic concept approach. In particular, a modularized and flexible management of taxonomic concept labels (TCLs) in conjunction with parent/child relationships and RCC–5 articulations – in some instances under multiple extensional or intensional readings (Section 8.II.1) – is out of scope for DwC. Certain aspects are covered by the TCS. However, this 2005-ratified standard needs revision and expansion, particularly in connection with a fully functional specimen data environment such as Symbiota.
We will adhere to DwC and TCS as much as is conducive to our representation needs. At the same time, this part of the project (Section 8.3.I: taxonomy/occurrence module expansion) is properly viewed as new work required for updating and expanding the TCS ("2.0"). Other services (e.g., GBIF, iDigBio) that 'just' manage DwC syntax and semantics, while not incompatible with our data, will nevertheless be unable to replicate our TCL-based specimen resolution services that critically require RCC–5 integration signals. As a stop-gap solution, we will provide links to alignments on GitHub and/or in DataOne in the "dynamicProperties" field.
At present Euler/X input and output data formats, including the input constraint .txt files and resulting .csv MIR files, are not covered by ratified standards (TDWG or other entities). However, both are ASCII-based, largely translatable into TCS terms and relationships, and easily manageable through standard control version systems (such as Git) that can automatically visualize version differences. The scale of this project – 2,000-3,000 alignments – presents an opportunity to create more formalized input and output data standards. The UIUC team will develop a simple alignment archive format (.aarc). We will also generate an associated and self-contained viewer tool to make taxonomy alignment products (i.e., input, output, and inference rules used to logically connect these products) transparent and reproducible.
Policies for Access and Sharing
Our project operates fully in the Public Domain. The Symbiota software code is published under the GNU General Public License (Version 3, June 2007), whereas the Euler/X code is published with the BSD license (also used by the Open Tree of Life project). All Symbiota-/SERNEC-held data and the new Euler/X alignments are published under the CC0 license (or similar, given certain collections records and image artefacts; see https://www.idigbio.org/content/idigbio-intellectual-property-policy; http://choosealicense.com/licenses/). UIUC's Ludäscher is a member of the DataONE Leadership Team and will work with colleagues in the DataONE Semantics and Provenance Working Group to explore sharing taxonomically (TCL) annotated datasets through DataONE.
Policies for Re-use and Distribution
Collection- and use case-based data will be published as Darwin Core Archive files. To disseminate DwC–A packages, we will use well-established and separate publication pathways from Symbiota to GBIF (http://www.gbif.org/dataset/) and iDigBio (https://www.idigbio.org/portal/publishers), as preferred by these aggregators. The transformed SERNEC portal will also publish our datasets, as DwC-A files and additionally using the expanded schema (syntax, semantics) for multi-TCL-to-specimen resolution that we will generate. This ensures that our use case results remain accessible and reproducible. Specific data packages authored in relation to the use case publications will be disseminated via means sanctioned by open access (option) journals, using repositories such as Dryad (http://datadryad.org/), figshare (https://figshare.com/), and Zenodo (http://zenodo.org/).
Plans for Archiving and Preservation
New software code will be published as releases through GitHub or similar openly accessible source code repositories (e.g., http://gitlab.com). SERNEC portal and use case data will be archived through redundant back-ups at ASU, in addition to GBIF and iDigBio. Data persistence will be further assured by establishing a new archival service relationship with DataONE, facilitated by Ludäscher, and specifically through addition of our Project data to the DataONE member node Knowledge Network for Biocomplexity (KNB) Data Respository (https://search.dataone.org/#profile/KNB).
Roles and Responsibilities
ASU (Franz, Gilbert) assume primary responsibility for project-based managing of data for Symbiota, SERNEC, and the Euler/X alignment repository on GitHub (https://github.com/taxonomic-concept-alignments). All Symbiota code (https://github.com/Symbiota) and contingent software for portal operation are open source. For select code testing purposes, ASU maintains an experimental portal on an institutionally supported VM server (http://hasbrouck.asu.edu/sandbox/). However, all actual SERNEC data are hosted only and directly by the NSF-supported iDigBio infrastructure, which has dedicated Symbiota data servers for multiple hosted data portals. We commit to iDigBio's rules for collaboration, particularly with regards to creating and resolving globally unique specimen identifiers; see https://www.idigbio.org/content/collaborating-idigbio-grant-proposals. UIUC (Ludäscher) is responsible for maintaining the new Euler/X code on GitHub (https://github.com/EulerProject/).
Project description
This ABI Development proposal is concerned with building a culture that increases trust in aggregated biodiversity data. We show that the meanings of taxonomic names are a variable in this context that needs to be explicitly modeled and controlled for. We will build a novel, multi-taxonomy conflict resolution service into a herbarium portal, as a pioneering effort that can be applied and propagated more widely.
1. Taxonomic names are (ambiguous) taxonomic concept lineage identifiers
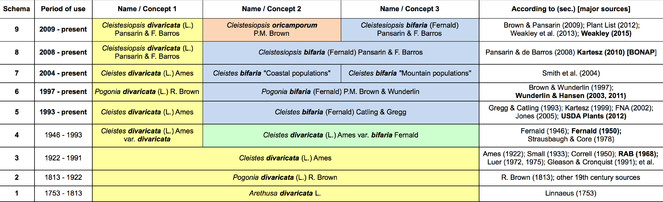
To motivate a complex theme – names, taxa, and concepts – we start with a concrete example. The species epithets "bifaria" (coined by
But here we should pause. The phrase "identifiers for species" could imply that we have converged on stable and accurate circumscriptions of two orchid species. It could even imply that we had 'gotten them right' since
Taxonomic concept labels and concept-to-concept articulations, represented in a tabular alignment of nine schemata, for the Cleistes use case (sec. A.S. Weakley). The vertical column position and width of taxonomic concept labels indicates taxonomic non-/congruence. The colors approximate taxonomic name lineages, e.g. blue for bifaria and yellow for divaricata.
Until 1946, divaricata had a wide taxonomic referent (= entity for which the name stands), whereas subsequently divaricata started to also stand for a narrower referent. Following
If names are potentially ambiguous, then how should we model the evolving relationships between identifiers, meanings, and natural entities? We propose the following definitions (
It follows that taxonomic names have three roles in our data systems (
The third role is critical for querying non-type specimens. Fig.
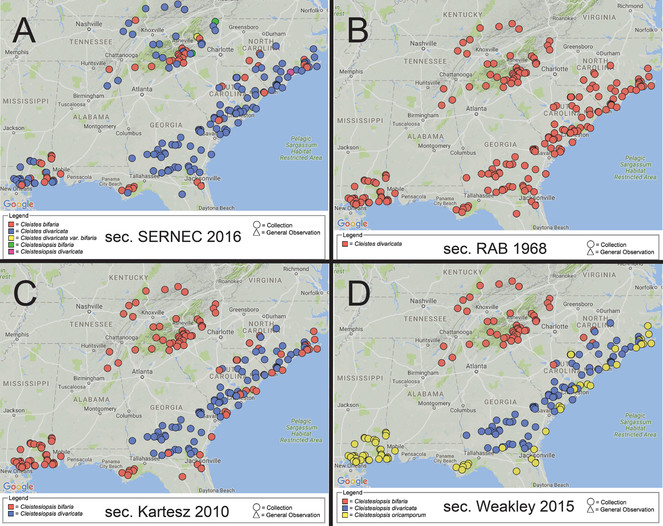
"Where do these endangered orchid species occur?" – visualizing the taxonomic variable for aggregated herbarium data. Mappings for the same 250 SERNEC specimens (not all resolved at this geographic scale) according to four distinct taxonomies. (A) sec.
Likely, one or another specification would lead the user to make distinct biological inferences based on these derivative maps. This is how the user can assess the robustness of their hypotheses vis-à-vis the taxonomic variable.
2. New syntax and semantics for identifying and articulating taxonomic concepts
What we describe is hard to do (
To begin building a solution, we need a new term for the identifier "Cleistesiopsis divaricata [name author, year] sec.
Thus, in addition to modeling TCLs, we need a new language to express concept-to-concept relations (
- Cleistesiopsis sec.
Weakley (2015) is a parent of Cleistesiopsis oricamporum sec.Weakley (2015)
Such parent/child relationships are explicit in the hierarchy asserted by the particular treatment. And the latter, between-hierarchies relationships are RCC–5 articulations, where "RCC" stands for Region Connection Calculus (
- Cleistesiopsis bifaria sec.
Weakley (2015) < Cleistesiopsis bifaria sec.Kartesz (2010) - Cleistesiopsis divaricata sec.
Weakley (2015) ! Cleistes divaricata var. bifaria sec.Fernald (1950) - Cleistesiopsis oricamporum sec.
Weakley (2015) == Cleistes bifaria "Coastal populations" sec.Smith et al. (2004)
Armed with the new syntax (TCLs) and semantics (parent/child relationships and RCC–5 articulations), we are much closer to responding to the counter-query "Please specify your preferred name usage".
3. Design and trust – a taxonomic 'synthesis' that nobody believes in
In Fig.
We need to be cautious in interpreting these 'mostly real' data visualizations that make SERNEC (2A) look dismal. The 250 specimens are housed in 33 different herbaria. They were vouchered over the period of 1869 to 2011, which likely means that they were variously re-/identified using any/all relevant treatments starting with
But this proposal is as much about the design of aggregating systems (trust) as it is about promoting more, and more accurate, identifications (quality). If we look at the
In particular, the system will not permit users to submit specimen queries in accordance with a particular taxonomic perspective, other than 'the portal consensus'. And we note that 'the consensus' is actually an evolving body of data, yet without adequate version tracking through time (
What needs to change? The prevailing name-based designs of aggregating systems improperly conflate two semi-independent processes. One might say with reasonable accuracy that, given a particular taxonomic perspective, the application of valid names and nomenclatural relationships is an undemocratic, logically contingent process. However, adherence to this or that perspective is democratic. At present, herbaria networked in
This proposal has conceptual, technical, social, and hence trust-related implications for biodiversity data science. The difference between the four visualizations (Fig.
4. Intellectual merit: Creating a trusted "big" biodiversity data culture
While our development focuses on
The service envisioned in Fig.
Below we describe why
5. Why SERNEC? Review of relevant prior work
A constellation of prior efforts in four different areas uniquely identifies SERNEC as the target for developing a concept-based system.
- SERNEC is an active, NSF-supported Thematic Collections Network (TCN), with currently 60 member herbaria (goal: 200) and nearly 1.97 million herbarium records (goal: 3–4 million). More than 85% of SERNEC's specimens are from the Mid-Atlantic and Southeastern United States region. Led by Zack Murrell (Appalachian State University), the community is primed to shift to taxonomic concept resolution.
- The
SERNEC (2016) portal and underlying data are sustained by the open source Symbiota software platform (Gries et al. 2014 ), hosted by the Integrated Digitized Biocollections project (iDigBio; see https://www.idigbio.org). Symbiota is the go-to solution for the majority of NSF-supported TCNs (August, 2016: 11/17 TCNs; ~ 25 portals; > 750 collections; > 32 million records; > 9 million images; > 7,300 users including 2,875 active contributors). It is the most impactful mid-level aggregator for millions of distributed, Darwin Core-compliant records (Wieczorek et al. 2012 ) that nevertheless allows full data access and authorship by individual collections and expert contributors. Co-PI (PI = Principal Investigator) Edward Gilbert (Arizona State University; ASU) is the lead Symbiota developer, and is singularly qualified to transform Symbiota as part of his developing biodiversity informatics research program. - Co-PI Bertram Ludäscher (University of Illinois Urbana-Champaign; UIUC) and PI Franz (ASU) are co-leaders of the Euler/X toolkit (
Chen et al. 2014b ,Dang et al. 2015 ,Ludäscher et al. 2016 ) for achieving logically consistent multi-taxonomy alignments (Section 6). Application of this toolkit will allow us to provide RCC–5 articulations at scale. Lastly, - Co-PI Alan Weakley (University of North Carolina; UNC) is the author of the Weakley Flora (
Weakley 2015 ; see http://www.herbarium.unc.edu/flora.htm and Fig.4 ). This 1320-page treatment covers nearly the entire SERNEC region, except southern Florida (expansion in progress). Weakley specifies as valid 7,000 taxonomic concepts at the sub-/specific level. In addition, he provides RCC–5 articulations to taxonomic concepts authored in multiple relevant preceding treatments (Fig.4 ; http://tinyurl.com/wf-rcc5). These logically actionable concept-to-concept reconciliation data are unique and invaluable to the project. We have on hand 75,621 RCC–5 articulations that reconcile each of Weakley's 7,000 concepts (on average nearly 11 times) with alternative concepts stemming from 465 sources, starting as early asCoulter and Rose (1900) and - and reaching the present time (
Schilling et al. 2015 ). The 20 most comprehensive treatments – e.g.Small (1933) ,Wunderlin and Hansen (2011) , orFNA (2015) – have 1,402–5,543 articulations, for a total of 66,996 or 88.6% of all articulations.
What do Weakley's RCC–5 articulations signal? Weakley's articulations measure the performance of taxonomic names as identifiers of taxonomic meanings (Table
Name-to-meaning reliability analysis of Weakley's RCC–5 data. Bold & italized font = reliable names (in pairwise alignments); regular font = name and/or meaning change; underlined font = totals.
|
Relationship (RCC–5 / names) |
== | > | < | >< | ? | Totals |
| Same name(s) | 43,185 | 625 | 1,540 | 15 | 24 | 45,389 |
| Different names | 13,836 | 6,433 | 9,000 | 228 | 735 | 30,232 |
| Totals | 57,021 | 7,058 | 10,540 | 243 | 759 | 75,621 |
Furthermore, Weakley's work focuses on providing one lowest level, closest matching articulation to a concept in another treatment. This has numerous implications. (1) Weakley's articulations do not directly address the genus level, although often species-level incongruences will propagate up (Fig.
6. Achieving scale with the Euler/X reasoning toolkit
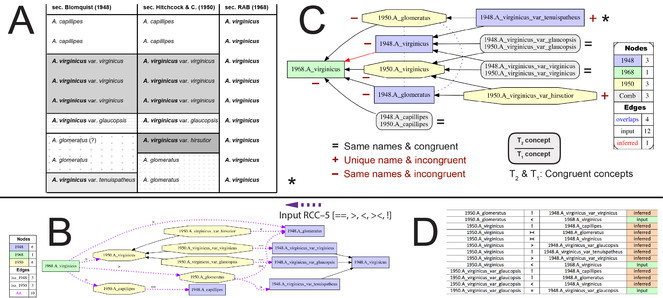
Application of Euler/X to data explicit and implicit in Weakley's Flora will yield 1.5–2.5 billion additional RCC–5 articulations. Here is how. The toolkit ingests two or more taxonomies (T1,T2,…,TN) at a time (
The input constraints and derived alignments can be visualized (Fig.
Euler/X toolkit products. (A) Part of the complex "Andropogon use case", with 1948/1950/1968 input sec.
Application of Euler/X will generate vast numbers of concept-to-concept relations that speak directly to the query: "To what extent can these two concepts be integrated?" The toolkit will create a comprehensive corpus of RCC–5 signals that will newly drive name-based integration for SERNEC specimen data.
7. ABI Development objectives: Taxonomic concept resolution for SERNEC
We target the ABI Development level because our innovations consist primarily of key increments to well-established service components, and in making the newly integrated infrastructure work in conjunction with SERNEC's specimen data.
- Through annual workshops and continuous research on specific use cases, we will foster community engagement to transform SERNEC into the first concept-based herbarium specimen data culture.
- We will apply the Euler/X reasoning toolkit to generate comprehensive genus- to variety-level concept alignments for at least 10 major treatments with relevance to SERNEC. Both visual and MIR-based toolkit products (> 109 RCC–5 articulations) will be stored in a GitHub repository for user access from the portal, and the RCC–5 articulations will drive the future reconciliation of specimen data.
- We will expand and optimize the Symbiota taxonomy and occurrences modules, and critical related user interfaces, to represent both name- and concept-based information. We will build modules to manage multiple concept taxonomies, utilize Euler/X reasoning products for specimen queries and visualizations, and perform semi-automated upgrades of identifications to entirely transition to TCLs.
- Using Symbiota's new identification module and the expertise of the UNC team and SERNEC collaborators, we will augment minimally 80% of the in-region specimen identifications (up to 1.6 million) from the current name-level to the most granular available TCL. Where needed, the identifications (and hence specimens) will be flagged with an "uncertainty signal" (need for study).
- We will research and publish at least 12 use cases "on the impact of controlling for the taxonomic variable" that showcase the diverse scientific (evolution, ecology) and societal (global change, conservation) significance of further developing a more robust, concepts-to-specimens data culture.
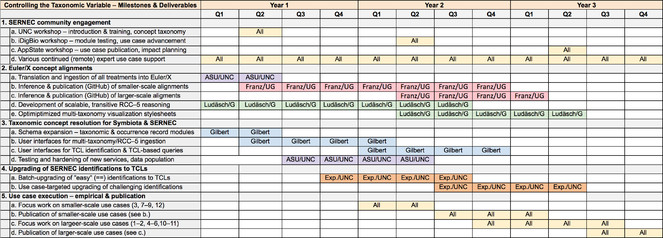
8. Research and implementation plan: Realizing objectives 1–5
8.1. Community engagement. Community engagement is absolutely critical because we aim to build a new data culture by example. Working with the
To further deepen the engagement, we have identified a core of 12 use cases that will be taken up from the planning to the publication stage by leaders within the SERNEC community (Section 8.5). To directly engage SERNEC scientists, we will hold annual workshops (2nd quarter of each project year) with as many as 10 non-local invitees plus 10-20 local participants at UNC (year 1), the University of Florida and iDigBio (year 2), and Appalachian State University (year 3). Workshop goals will evolve with the advancement of use cases. Each workshop will run for two full days, plus travel. During the interim periods, we will communicate virtually with use case groups (e.g. via iDigBio's Adobe Connect) and through monthly updates to the
8.2. Euler/X concept alignments. ASU and UIUC will concentrate on this task, with Franz mentoring undergraduate students at ASU to create and publish the alignments, and Ludäscher mentoring a graduate (Ph.D.) student at UIUC to develop new toolkit capabilities for special reasoning and visualization challenges of the SERNEC use case. Weakley's group (UNC) will provide expert input as needed.
I. Scope. We will produce comprehensive – all with all – alignments for the 6-12 most abundantly applied treatments for SERNEC, given the taxonomic subgroup (see
II. Feasibility. The task of producing two types of 413 family-level alignments that are 6-12 taxonomies deep and reciprocally comprehensive may appear daunting. We are certain that it is not, given prior experience, efforts, and project resources allocated to this task. The reasoning capacity is already there (
III. Approach. We have run thousands of successful alignments with Euler/X, including larger sub-alignments (all Gymnospermae, all Rosaceae) of
IV. New Euler/X development.
8.3. Adding taxonomic concept representation to Symbiota and SERNEC. This objective requires a large part of the project's resources for new Symbiota development at ASU. Three major task domains are involved. (1) Symbiota's underlying taxonomic and occurrence schemas must be expanded to support TCLs, source-specific parent/child relationships, and RCC–5 articulations. (2) A subset of Symbiota's graphic user interfaces will be changed accordingly, and new interfaces will be created to manage multiple taxonomies and efficiently upgrade specimen identifications to TCLs. (3) A name-to-concept transition plan for the SERNEC portal will be executed such that (a) existing named-based data are not functionally compromised and (b) new concept-based data become the portal norm – most immediately to support our use cases. Below we describe the sequence of actions that will achieve this transition.
I. Schema expansion. The expansion of Symbiota's taxonomic and occurrence modules will be guided by the remarkable example of Avibase (
II. User interfaces. We will upgrade a critical subset of Symbiota's interfaces to enable concept use. In particular, we will modify the taxonomy viewing and editing interfaces to only accept the new syntax and semantics. New concept taxonomies can be uploaded piece-meal or through batch functions. We will also create simple formatting and loading tools to ingest multiple taxonomies into the Euler/X alignment toolkit and re-integrate the outcomes (MIR) into the RCC–5 table. Based on the latter, we will generate a new "incongruence alert" table that entails precisely those taxonomic names that, when searched by users, require additional specification of a TCL to identify a consistent circumscription (Figs
Symbiota already has an effective visualization interface for single taxonomies. Rather than building a new multi-tree visualization interface – which is both difficult and redundant (
We will reconfigure the occurrence identification interface to interact with the new taxonomy module. Again, this will include drop-down options to select preferred sources, view alignments, and populate a TCL. Very substantive upgrades will be made to the add batch determinations interface, which presently permits selecting names or individual specimens. In collaboration with SERNEC experts, we will expand this interface to represent the subset of Darwin Core fields most decisive for filtering occurrences so that batch updates can follow. Target fields will include (e.g.) the source collection, collecting locality and date, collector/identifier information (who/when?), and references (where available). Combinations of these fields will facilitate smart queries of the kind: "show me all specimens in this region, collected in this time period, and identified to this name by members of this herbarium community". These queries, combined with expert knowledge and specimen images, will facilitate upgrades of many identifications to TCLs at once. A second, innovative map-based determinations function will be developed as an extension of Symbiota's Map Search module. It will allow experts to gather specimen sets for batch determinations directly through the map interface (http://tinyurl.com/sernec-mapint), by using area shape selectors. Because granular taxonomic concepts are often geographically separated (Fig.
Lastly, we will transform the primary search and display specimens interfaces. Key goals are to promote TCL-based specimen queries and mappings, with the option to relabel an initially queried set according to an alternative treatment (Fig.
III. Transition plan. Realizing the above changes requires a sound transition plan. It is critical not to break existing services while building new ones for transition. We also recall that 57.1% of the RCC–5 articulations identify reliable name usages that (at present) need no additional specification (Table
Once the taxonomy module is expanded, it is necessary to 'reify' the SERNEC consensus taxonomy (Fig.
8.4. Augmenting SERNEC specimen identifications to TCLs. Using the new tools, our goal is to upgrade minimally 80% of all specimen identifications from "sec.
Overview of 12 use cases. Headers: valid names sec.
| # |
Names sec. |
Taxonomic diversity sec. |
Specimens in |
Names in |
Reliability ratio | Impact | Use case lead |
| 1 | Andropogon "complex" | 7 species | 4 varieties | 2,696 | 16 | 14 : 90 (13.5%) | Dis - Div - Evo | Weakley |
| 2 | Asarum & Hexastylis | 14 species | 5 varieties | 3,564 | 36 | 87 : 110 (44.2%) | Dis - Div - Phy | Murrell |
| 3 | Cleistes & Cleistesiopsis | 3 species | 250 | 12 | 8 : 47 (14.6%) | Con - Dis - Phy | Weakley |
| 4 | Coreopsis | 23 species | 11 varieties | 4,561 | 56 | 185 : 155 (54.4%) | Eco - Evo - HBG | Weakley |
| 5 | Cornus | 11 species | 5,575 | 40 | 104 : 63 (62.2%) | Eco - Evo - HBG | Murrell |
| 6 | Euphorbia | 50 species | 5,747 | 190 | 247 : 213 (53.7%) | Con - Dis - Eco | Alford |
| 7 | Galactia | 7 species | 1 variety | 1,408 | 23 | 61 : 49 (55.5%) | Dis - Div - Eco | Franck |
| 8 | Gonolobus & Matelea | 9 species | 2 varieties | 1,571 | 28 | 48 : 43 (52.7%) | Eco - Evo - Phy | Fishbein |
| 9 | Lantana | 4 species | 1 variety | 659 | 22 | 22 : 19 (53.6%) | Eco - Exo - GCB | Franck |
| 10 | Liatris | 28 species | 4 varieties | 4,200 | 70 | 121 : 185 (39.6%) | Con - Evo - Pol | Alford |
| 11 | Magnolia | 9 species | 4 varieties | 5,135 | 45 | 46 : 114 (28.8%) | Cul - Evo - HBG | Weakley |
| 12 | Monotropis | 2 species | 56 | 4 | 13 : 19 (40.6%) | Eco - Evo - GCB | Weakley |
Improving identifications will be facilitated by technology, but is only feasible because of the direct involvement of experts. Some 15-30% of the SERNEC records have partial identification-related information recorded (expert, year, taxonomic reference used). We will utilize these data to identify the best-fitting TCL. Collective experience strongly indicates that, even for problematic cases, a remotely working expert can confidently assert TCL identifications by drawing on their sophisticated background knowledge of spatially/temporally localized identification practices. For instance, a very large number of non-Floridian SERNEC herbaria have treated
Thousands of herbarium sheets will nevertheless remain "uncertain" with regards to the narrowest TCL. Removing uncertainty may require direct study of morphological/molecular data, likely in the context of new revisions. Although hypotheses are weakened in such cases, we regard this as a positive contribution to explicitly identify 'problem specimens'. In analogy to the "alert" table for incongruent name usages, we will create a special flag for uncertain TCL identifications. Flagged specimens will be retrievable by query, and uniquely colored on maps, with an option to show only non-ambiguous specimens.
8.5. Use case selection, approach, and impact. We have enlisted five SERNEC botanists (plus the UNC postdoc; Section 9) to lead 12 use cases (Table
I. Use case particulars. The Andropogon glomeratus-virginicus "complex" is notorious for its taxonomic instability (
Euphorbia (spurge family) is the most speciose complex, including recent introductions not yet keyed out by
II. Research approach. We expect that use case leaders will engage many additional SERNEC members. Although the TCL identification efforts (Section 8.4) will be similar in each case, the ultimate research goals will vary greatly. Some may take on the form of a review – though rooted in specimen-level data and visualizations – of taxonomic inconsistencies affecting our basic understanding of biodiversity and distribution. Others may reassess the specimen-level evidence and inter-taxonomic robustness of very specific ecological or evolutionary hypotheses. Still others may characterize to what extent conservation and global change assessments are contingent on a specific taxonomic commitment (
III. Innovative impact. Rather than specifying each worthwhile research question, we exemplify the kinds of questions that our development will facilitate, and why this matters. Accordingly, in the case of Andropogon, users can query (often sequentially):
- "Show me all specimens identified to the taxonomic name Andropogon virginicus in the Carolinas" [returns many records, resolved only with the ambiguity of an incongruent taxonomic concept lineage].
- "Now show me all specimens identified to the TCL Andropogon virginicus sec.
Weakley 2015 " [returns a subset of these records, reflecting a choice for this particular, granular TCL resolution]. - "Now me all specimens of Andropogon virginicus sec.
Radford et al. 1968 , yet translated into the TCLs sec.FNA 2015 " [returns (again) many records, but specifically represents and contrasts two treatments, as opposed to providing the ambiguous lineage view of (1)]. - "Show me all specimens whose identifications are ambiguous with regards to
FNA 2015 versusWeakley 2015 " [using the "uncertainty" flag, points to specimens needing further study]. - "For the Carolinas and for this inclusive specimen set, show me the composite least versus most granular taxonomic perspective(s) available" [returns a potentially multi-taxonomy 'composite' that represents opposite extremes in resolution granularity].
- "Save and output all results in Darwin Core Archive format".
This is what we mean by "controlling the taxonomic variable". The services will be basic, as dictated by realism, and the control offered to users is not explicitly of a statistical kind. Yet we are confident that queries (2-6) – which are not supported by any existing herbarium specimen network – will yield impactful outcomes when applied to the aforementioned use cases and research goals. We will work to carry each of these to publication in international journals with open access options that variously expand the reach of our approach, such as Biodiversity Data Journal, Conservation Biology, Global Ecology and Biogeography, PeerJ, PLoS ONE, Systematic Biology, Taxon, and Trends in Ecology and Evolution.
9. Lead personnel and management
The project lead personnel – Franz, Gilbert, Ludäscher, and Weakley – is introduced in Section 5. Franz will mentor 15 undergraduate students at ASU to achieve the Euler/X alignments (Section 8.2.I–III). At UIUC, Ludäscher will mentor a Ph.D. student who will concentrate on the reasoning and visualization challenges (Section 8.2.IV). At UNC, Weakley will mentor a postdoc (year 1), and utilize applications analyst Michael Lee to provide critical support at the interface of Symbiota module development, data population, and service optimization for the use cases. Weakley and the postdoc will play an immense role in overseeing the rapid integration of the floristic legacy, new tools, and expert contributions. Gilbert and Lee will translate new conceptual and TCL identification-related functions and data from various sources into the transformed SERNEC portal (Sections 8.3 & 8.4). We invest significant resources to support expert co-leadership of the use cases (Sections 8.1 & 8.5).
Fig.
10. Broader impacts – scientific and educational
I. Scientific. The intellectual case was presented in Sections 1–4. Our project is focused on just one herbarium network, but the potential impact is much wider. Space does not permit reviewing the many aggregators that concede, in one form or another, that taxonomic concept resolution is needed but seemingly out of reach. The list includes (e.g.) Catalogue of Life, GenBank, Global Biodiversity Information Facility, Global Names Architecture, iDigBio, Open Tree of Life, and the Taxonomic Name Resolution Service (
This project is designed to advance a global agenda, by demonstrating that conceptual and technical challenges can be addressed at scale if communities are willing to engage in concept taxonomy. Trust in data is also a design feature of allowing expert access and resolving multiple conflicting views that reflect the realities of ongoing taxonomic research. If only 1% of SERNEC's data display the issues shown in Fig.
II. Educational. We will directly train one postdoctoral researcher (UNC), two Ph.D. students (ASU, UIUC), and at least 15 undergraduate students (ASU). Each of our workshops will capacitate 10-15 SERNEC experts, who in turn can inform and recruit colleagues and students at their home herbaria. Project members Alford, Fishbein, Franck, Franz, Gilbert, Murell, Soltis, and Weakley regularly offer plant/biodiversity courses to undergraduate students at their respective institutions, reaching an estimated 300-500 students per year, with ca. 10-40% minority students (range: Oklahoma – Mississippi). Each has committed to integrating our project's theme and use cases as new sections into their future biodiversity teaching plans. At ASU, this will include two new three-hour sections of the undergraduate-focused biodiversity informatics course "Discovering Biodiversity – Field to Database", offered in the spring of 2017 and 2019 to 25 students. At each institution, project members will make a sustained, systematic effort to recruit new students from underrepresented groups, working through institutional (e.g., sponsored STEM minority mentor programs) and local student organizations to advertise project opportunities and thereby proactively broaden participation.
Murrell's leadership of SERNEC will promote our advances with nearly 200 herbarium scientists in the region. Alford's involvement in the Magnolia grandiFLORA project (http://www.mississippiplants.org/), which has an educational component for K–12 teachers, will add exposure. At ASU, Franz and Gilbert will promote the project through virtual and personal outreach, aided by their leadership of the Biodiversity Knowledge Integration Center (BioKIC; https://biokic.asu.edu/). We will publish a BioKIC monthly blog post with project updates, to be shared with the iDigBio/Symbiota Working Group. Conference presentations will mainly target the global TDWG community (http://www.tdwg.org/).
We budget funds for two additional forms of outreach. The first will be a feature story "Where do plant species occur?" (see Fig.
11. Sustainability
Mid-term prospects (~ 5-15 years) for our development and data innovations are very strong. Our project operates inside an upward-trending information culture (
Results from prior NSF support
Details are not provided here; however, the following NSF-funded projects were reviewed (intellectual merit, broader impacts) for each Co-/PI. This information is publicly available through the NSF website (links provided here).
- PI Franz. NSF DEB-1155984. CAREER: Systematics of eustyline and geonemine weevils: connecting and contrasting Caribbean and Neotropical mainland radiations. http://nsf.gov/awardsearch/showAward?AWD_ID=1155984
- Co-PI Gilbert. NSF-DBI 0743827. Symbiota, a virtual flora model for the Southwestern United States. http://www.nsf.gov/awardsearch/showAward?AWD_ID=0743827
- Co-PI Ludäscher. NSF IIS-1118088. III: Small: A logic-based, provenance-aware system for merging scientific data under context and classification constraints. http://nsf.gov/awardsearch/showAward?AWD_ID=1118088
- Co-PI Weakley. NSF-DBI 1410439. Digitization TCN: Collaborative Research: The key to the cabinets: building and sustaining a research database for a global biodiversity hotspot. http://www.nsf.gov/awardsearch/showAward?AWD_ID=1410439
Acknowledgements
The authors acknowledge critical proposal and input by Mac Alford, Mark Fishbein, Alan Franck, Michael Lee, Zack Murrell, and Pamela Soltis. This acknowledgment need not imply that the collaborators are responsible for the overall goals and content of the proposal.
Funding program
National Science Foundation (U.S.A.): Division of Biological Infrastructure: Advances in Biological Informatics.
Grant title
Collaborative Research: ABI Development: Controlling the taxonomic variable: Taxonomic concept resolution for a southeastern herbarium portal.
Hosting institution
Arizona State University, with the University of Illinois Urbana-Champaign and the University of North Carolina collaborating.
Ethics and security
None apparent.
Author contributions
NMF had primary content-related and organizational responsibilities, however all authors contributed (variously) to most proposal aspects. ASW contributed the RCC–5 data upon which much of the concept alignment objectives are based.
Conflicts of interest
None apparent.
References
- Orchidaceae: Illustrations and Studies of the Family Orchidaceae, Issuing from the Ames Botanical Laboratory, Fascile 7.Ames Botanical Laboratory,North Easton, Massachusetts,174pp. https://doi.org/10.5962/bhl.title.15433
- Molecular phylogeny of the Magnoliaceae: the biogeography of tropical and temperate disjunctions.American Journal of Botany88(12):2275‑2285. https://doi.org/10.2307/3558389
- Intraspecific sequence variation of cpDNA shows two distinct groups within Magnolia virginiana L. of Eastern North America and Cuba.Castanea76(1):118‑123. https://doi.org/10.2179/10-018.1
- The concept of "potential taxa" in databases.Taxon44(2):207‑212. https://doi.org/10.2307/1222443
- Networking taxonomic concepts – uniting without 'unitary-ism'; pp. 13-22.Biodiversity Databases: from Cottage Industry to Industrial Networks. Curry, G., Humphries, C. (Eds.). Systematics Association Special Volume 73. Taylor & Francis, Boca Raton. https://doi.org/10.1201/9781439832547.ch3
- An integrative and dynamic approach for monographing species-rich plant groups – building the global synthesis of the angiosperm order Caryophyllales.Perspectives in Plant Ecology, Evolution and Systematics17(4):284‑300. https://doi.org/10.1016/j.ppees.2015.05.003
- Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology.AMBIO: A Journal of the Human Environment37(2):114‑118. https://doi.org/10.1579/0044-7447(2008)37[114:ecitbs]2.0.co;2
- Provenance in collection-oriented scientific workflows.Concurrency and Computation: Practice and Experience20(5):519‑529. https://doi.org/10.1002/cpe.1226
- The taxonomic name resolution service: an online tool for automated standardization of plant names.BMC Bioinformatics14(1):16. https://doi.org/10.1186/1471-2105-14-16
- Pogonia, p. 203.Hortus Kewensis; or, a Catalogue of the Plants Cultivated in the Royal Botanic Garden at Kew, Second Edition, Volume 5.Longman, Hurst, Rees, Orme, and Brown,London,568pp. https://doi.org/10.5962/bhl.title.105339
- Systematics of the Andropogon virginicus complex (Gramineae).Journal of the Arnold Arboretum64(2):171‑254. https://doi.org/10.5962/bhl.part.27406
- Provenance in databases: why, how, and where.Foundations and Trends in Databases1(4):379‑474. https://doi.org/10.1561/1900000006
- A hybrid diagnosis approach combining Black-Box and White-Box reasoning; 127-141.Rules on the Web. From Theory to Applications. Proceedings of the 8th International Symposium, RuleML 2014, Co-located with the 21st European Conference on Artificial Intelligence, ECAI 2014, Prague, Czech Republic, August 18-20, 2014. Lecture Notes in Computer Science 8620. https://doi.org/10.1007/978-3-319-09870-8_9
- Euler/X: a toolkit for logic-based taxonomy integration; arXiv:1402.1992 [cs.LO].arXiv2014:1‑8. URL: http://arxiv.org/abs/1402.1992
- Provenance for explaining taxonomy alignments; pp. 258-260.Provenance and Annotation of Data and Processes. Revised Selected Papers of the 5th International Provenance and Annotation Workshop, IPAW 2014, Cologne, Germany, June 9-13, 2014. Lecture Notes in Computer Science 8628. https://doi.org/10.1007/978-3-319-16462-5_27
- Monograph of the North American Umbelliferae.Contributions of the United States National Herbarium7(1):1‑256. https://doi.org/10.5962/bhl.title.38223
- Phylogeny of Eastern North American Coreopsis (Asteraceae-Coreopsideae): insights from nuclear and plastid sequences, and comments on character evolution.American Journal of Botany92(2):330‑336. https://doi.org/10.3732/ajb.92.2.330
- ProvenanceMatrix: a visualization tool for multi-taxonomy alignments.CEUR Workshop Proceedings1456:13‑24. URL: http://ceur-ws.org/Vol-1456/paper2.pdf
- i4Life - Indexing for Life. D6.1. Integrated access to CoL in GBIF, Workpackage 6, pp. 1-4.Capacities Programme of Framework 7: EC e-Infrastructure Programme - Virtual Research Communities - INFRA-2010-2.4pp. URL: http://www.i4life.eu/i4lifewebsite/wp-content/uploads/2012/12/D6-1-Integrated-Access-to-CoL-in-GBIF_V4.pdf
- GBIF Checklist Bank and the Backbone Taxonomy. http://www.slideshare.net/mdoering/checklist-bank-and-backbone. Accessed on: 2016-8-15.
- Updating the GBIF Backbone. http://gbif.blogspot.co.uk/2016/04/updating-gbif-backbone.html. Accessed on: 2016-8-15.
- Phylogenetics, morphological evolution, and classification of Euphorbia subgenus Euphorbia.Taxon62(2):291‑315. https://doi.org/10.12705/622.1
- Proposed rules for the incorporation of nomina of higher-ranked zoological taxa in the International Code of Zoological Nomenclature. 1. Some general questions, concepts and terms of biological nomenclature.Zoosystema27(2):365‑426. URL: http://sciencepress.mnhn.fr/sites/default/files/articles/pdf/z2005n2a8.pdf
- Changes in Galactia (Fabaceae) of the southeastern United States.SIDA, Contributions to Botany8(2):170‑180. URL: http://www.jstor.org/stable/23909679
- Evolutionary prediction of medicinal properties in the genus Euphorbia L.Scientific Reports6:30531. https://doi.org/10.1038/srep30531
- The NCBI Taxonomy database.Nucleic Acids Research40:D136‑D143. https://doi.org/10.1093/nar/gkr1178
- Some orchids of the Manual range.Rhodora48(572):184‑197. URL: http://www.jstor.org/stable/23304761
- Some orchids of the Manual range.Rodora48(571):161‑162. URL: http://www.jstor.org/stable/i23303797
- Gray's Manual of Botany, Eigth (Centennial) Edition.American Book Company,New York,1632pp.
- Notes on Magnoliaceae IV.Blumea - Biodiversity, Evolution and Biogeography of Plants49(1):87‑100. https://doi.org/10.3767/000651904x486214
- Conservation of taxonomically difficult species: the case of the Australian orchid, Microtis angusii.Conservation Genetics7(6):847‑859. https://doi.org/10.1007/s10592-006-9119-8
- Flora of North America Editorial Committee, eds. 1993+. Flora of North America North of Mexico. 16+ vols. New York and Oxford. Vol. 1, 1993; vol. 2, 1993; vol. 3, 1997; vol. 4, 2003; vol. 5, 2005; vol. 7, 2010; vol. 8, 2009; vol. 19, 2006; vol. 20, 2006; vol. 21, 2006; vol. 22, 2000; vol. 23, 2002; vol. 24, 2007; vol. 25, 2003; vol. 26, 2002; vol. 27, 2007; vol 28, 2014; vol. 9, 2014; vol. 6, 2015.http://floranorthamerica.org. Accessed on: 2016-8-15.
- Big data for forecasting the impacts of global change on plant communities.Global Ecology and Biogeography2016:1‑12. https://doi.org/10.1111/geb.12501
- Towards a language for mapping relationships among taxonomic concepts.Systematics and Biodiversity7(1):5‑20. https://doi.org/10.1017/s147720000800282x
- Taxonomy - for computers.biorXiv2015:1‑19. https://doi.org/10.1101/022145
- Biological taxonomy and ontology development: scope and limitations.Biodiversity Informatics7(1):45‑66. https://doi.org/10.17161/bi.v7i1.3927
- On the use of taxonomic concepts in support of biodiversity research and taxonomy, pp. 63-86.The New Taxonomy, Systematics Association Special Volume, Series 74.Taylor & Francis,Boca Raton. https://doi.org/10.1201/9781420008562.ch5
- Reasoning over taxonomic change: exploring alignments for the Perelleschus use case.PLoS ONE10(2):e0118247. https://doi.org/10.1371/journal.pone.0118247
- Names are not good enough: reasoning over taxonomic change in the Andropogon complex.Semantic Web (IOS)7:1‑23. https://doi.org/10.3233/SW-160220
- Two influential primate classifications logically aligned.Systematic Biology65(4):561‑582. https://doi.org/10.1093/sysbio/syw023
- An open graph visualization system and its applications to software engineering.Software: Practice and Experience30(11):1203‑1233. https://doi.org/10.1002/1097-024x(200009)30:113.3.co;2-e
- Global Biodiversity Information Facility. Darwin Core Archives – How-To Guide, Version 1, Released on 1 March 2011 (contributed by Remsen, D., K. Braak, M. Döring & T. Robertson), Copenhagen. http://links.gbif.org/gbif_dwca_how_to_guide_v1. Accessed on: 2016-8-15.
- The concept problem in taxonomy: importance, components, approaches.Schriftenreihe für Vegetationskunde39:4‑15. URL: http://www.nhbs.com/series/64263/schriftenreihe-fur-vegetationskunde
- Assembling and navigating the potential taxon graph.Schriftenreihe für Vegetationskunde39:71‑82. URL: http://www.nhbs.com/series/64263/schriftenreihe-fur-vegetationskunde
- A survey of multiple tree visualisation.Information Visualization9(4):235‑252. https://doi.org/10.1057/ivs.2009.29
- Comparison of size vs. life-state classification in demographic models for the terrestrial orchid Cleistes bifaria.Biological Conservation129(1):50‑58. https://doi.org/10.1016/j.biocon.2005.09.044
- Symbiota – a virtual platform for creating voucher-based biodiversity information communities.Biodiversity Data Journal2:e1114. https://doi.org/10.3897/bdj.2.e1114
- Synthesis of phylogeny and taxonomy into a comprehensive tree of life.Proceedings of the National Academy of Sciences112(41):12764‑12769. https://doi.org/10.1073/pnas.1423041112
- Red List assessments of East African chameleons: a case study of why we need experts.Oryx49(4):652‑658. https://doi.org/10.1017/s0030605313001427
- Native and Naturalized Leguminosae (Fabaceae) of the United States (Exclusive of Alaska and Hawaii).Monte L. Bean Life Science Museum, Brigham Young University,Provo, Utah,1007pp. [ISBN978-0842523967]
- Phylogenetic revision of Minyomerus Horn, 1876 sec. Jansen & Franz, 2015 (Coleoptera, Curculionidae) using taxonomic concept annotations and alignments.ZooKeys528:1‑133. https://doi.org/10.3897/zookeys.528.6001
- Identifying and relating biological concepts in the Catalogue of Life.Journal of Biomedical Semantics2(1):7. https://doi.org/10.1186/2041-1480-2-7
- Trends in access of plant biodiversity data revealed by Google Analytics.Biodiversity Data Journal2:e1558. https://doi.org/10.3897/bdj.2.e1558
- Floristic Synthesis of North America, Version 9-15-2010. Biota of North America Program (BONAP), Chapel Hill. http://www.bonap.org/. Accessed on: 2016-8-15.
- Phylogenetic relationships in Asarum (Aristolochiaceae) based on morphology and ITS sequences.American Journal of Botany85(10):1454‑1467. https://doi.org/10.2307/2446402
- Scientific names are ambiguous as identifiers for biological taxa: their context and definition are required for accurate data integration, pp. 80-95.Data Integration in the Life Sciences: Proceedings of the Second International Workshop, San Diego, DILS 2005, LNBI 3615. https://doi.org/10.1007/11530084_8
- ITS sequences and phylogenetic relationships in Bidens and Coreopsis (Asteraceae).Systematic Botany24(3):480. https://doi.org/10.2307/2419701
- Comparative analysis of the reproductive ecology of Monotropa and Monotropsis: two mycoheterotrophic genera in the Monotropoideae (Ericaceae).American Journal of Botany96(7):1337‑1347. https://doi.org/10.3732/ajb.0800319
- Referenzliste der Moose Deutschlands.Schriftenreihe für Vegetationskunde35:1‑519. URL: https://www.amazon.de/Referenzliste-Moose-Deutschlands-Schriftenreihe-Vegetationskunde/dp/3784335047
- Synopsis of Gonolobus s. l. (Apocynaceae, Asclepiadoideae) in the United States and its territories, including lectotypification of Lachnostoma arizonicum.Harvard Papers in Botany13(2):209‑218. https://doi.org/10.3100/1043-4534-13.2.209
- Managing invasive plants on public conservation forestlands: application of a bio-economic model.Forest Policy and Economics11(4):237‑243. https://doi.org/10.1016/j.forpol.2009.03.004
- Classificatory theory in biology.Biological Theory7(4):338‑345. https://doi.org/10.1007/s13752-012-0049-z
- Avibase – a database system for managing and organizing taxonomic concepts.ZooKeys420:117‑135. https://doi.org/10.3897/zookeys.420.7089
- Species plantarum, exhibentes plantas rite cognitas, ad genera relatas, cum differentiis specificis, nominibus trivialibus, synonymis selectis, locis natalibus, secundum systema sexuale digestas. Tomus 1.Laurentii Salvii,Holmiae,560pp. https://doi.org/10.5962/bhl.title.669
- Systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis, Editio 10.Laurentii Salvii,Holmiae,824pp. https://doi.org/10.5962/bhl.title.542
- Reproductive biology and breeding system of Gonolobus suberosus (Asclepiadaceae).Journal of the Torrey Botanical Society125(3):183‑193. https://doi.org/10.2307/2997216
- Euler Project – Reasoning Over Taxonomies. Release date:2016-8-01. URL: https://github.com/EulerProject
- Estimating species diversity and distribution in the era of Big Data: to what extent can we trust public databases?Global Ecology and Biogeography24(8):973‑984. https://doi.org/10.1111/geb.12326
- The varieties of Liatris elegans (Asteraceae).SIDA, Contributions to Botany20(2):597‑603. URL: http://www.jstor.org/stable/41968077
- The vertebrate taxonomy ontology: a framework for reasoning across model organism and species phenotypes.Journal of Biomedical Semantics4(1):34. https://doi.org/10.1186/2041-1480-4-34
- Phylogenetic relationships in Cornus (Cornaceae).Systematic Botany18(3):469. https://doi.org/10.2307/2419420
- Examination of species boundaries in Hexastylis contracta Blomquist and H. rhombiformis Gaddy. (Abstract.).American Journal of Botany (Supplement)85:146‑147.
- Digitization workflows for flat sheets and packets of plants, algae, and fungi.Applications in Plant Sciences3(9):1500065. https://doi.org/10.3732/apps.1500065
- The Meaning of Meaning: a Study of the Influence of Language upon Thought and of the Science of Symbolism. 1st Edition.Kegan Paul,London,396pp. [ISBN978-0156584463]
- When integration fails: prokaryote phylogeny and the tree of life.Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences44(4):551‑562. https://doi.org/10.1016/j.shpsc.2012.10.003
- An Edit script for taxonomic classifications.BMC Bioinformatics6(1):208. https://doi.org/10.1186/1471-2105-6-208
- Taxonomic notes on Pogonieae (Orchidaceae): Cleistesiopsis, a new genus segregated from Cleistes, and description of two new South American species, Cleistes batistana and C. elongata.Kew Bulletin63(3):441‑448. https://doi.org/10.1007/s12225-008-9047-5
- A new genus for the North American Cleistes.North American Native Orchid Journal15(1):50‑58. URL: https://www.scribd.com/document/42865188/March-2009-North-American-Native-Orchid-Journal
- Monograph of Euphorbia sect. Tithymalopsis (Euphorbiaceae).Edinburgh Journal of Botany55(2):161‑208. https://doi.org/10.1017/s0960428600002122
- Challenges with using names to link digital biodiversity information.Biodiversity Data Journal4:e8080. https://doi.org/10.3897/bdj.4.e8080
- VegBank – a permanent, open-access archive for vegetation-plot data.Biodiversity & Ecology4:233‑241. https://doi.org/10.7809/b-e.00080
- The Essential Peirce, Volume 2. Peirce Edition Project (Editor).Indiana University Press,Bloomington,624pp. URL: http://www.iupress.indiana.edu/product_info.php?products_id=21333 [ISBN978-0-253-21190-3]
- Alternate species concepts as bases for determining priority conservation areas.Conservation Biology13(2):427‑431. https://doi.org/10.1046/j.1523-1739.1999.013002427.x
- A "taxonomic affidavit": why it is needed?Integrative Zoology2(2):57‑59. https://doi.org/10.1111/j.1749-4877.2007.00044.x
- Manual of the Vascular Flora of the Carolinas.University of North Carolina Press,Chapel Hill,1245pp. URL: http://uncpress.unc.edu/books/T-766.html [ISBN978-0-8078-1087-3]
- A spatial logic based on regions and connection, pp. 165-176.Proceedings of the Third International Conference on the Principles of Knowledge Representation and Reasoning.Morgan Kaufmann,Los Altos.
- The use and limits of scientific names in biological informatics.ZooKeys550:207‑223. https://doi.org/10.3897/zookeys.550.9546
- Cryptic and overlooked: species delimitation in the mycoheterotrophic Monotropsis (Ericaceae: Monotropoideae).Systematic Botany39(2):578‑593. https://doi.org/10.1600/036364414x680762
- Contextual cross-referencing of species names for fiddler crabs (genus Uca): an experiment in cyber-taxonomy.PLoS ONE9(7):e101704. https://doi.org/10.1371/journal.pone.0101704
- Primate taxonomy: species and conservation.Evolutionary Anthropology: Issues, News, and Reviews23(1):8‑10. https://doi.org/10.1002/evan.21387
- Taxonomy of Lantana sect. Lantana (Verbenaceae): I. Correct application of Lantana camara and associated names.SIDA, Contributions to Botany22(1):381‑421. URL: http://www.jstor.org/stable/41968588
- Taxonomy of Lantana sect. Lantana (Verbenaceae): II. Taxonomic revision.Journal of the Botanical Research Institute of Texas6(2):403‑441. URL: http://www.jstor.org/stable/41972430
- Barcoding the Asteraceae of Tennessee, tribe Cichorieae.Phytoneuron2019(19):1‑8. URL: http://www.phytoneuron.net/2015Phytoneuron/19PhytoN-TennCichorieae.pdf
- Unitary or unified taxonomy?Philosophical Transactions of the Royal Society B: Biological Sciences359(1444):699‑710. https://doi.org/10.1098/rstb.2003.1456
- SouthEast Regional Network of Expertise and Collections, On-Line Portal; Community website available at http://sernec.appstate.edu. http://sernecportal.org. Accessed on: 2016-8-15.
- A new system for the family Magnoliaceae; pp. 55-71.Proceedings of the Second International Symposium on the Family Magnoliaceae; Nianhe, X., Qingwen, Z., Fengxia, X., Qigen, W. (Eds.),Xia Nianhe, Zeng Qingwen, Xu Fengxia, Wu Qigen.304pp. [ISBN9787560973494].
- Manual of the Southeastern Flora: Being Descriptions of the Seed Plants Growing Naturally in Florida, Alabama, Mississippi, Eastern Louisiana, Tennessee, North Carolina, South Carolina and Georgia.J.K. Small,New York,1554pp. https://doi.org/10.5962/bhl.title.696
- Genetic discontinuities among populations of Cleistes (Orchidaceae, Vanilloideae) in North America.Botanical Journal of the Linnean Society145(1):87‑95. https://doi.org/10.1111/j.1095-8339.2003.00265.x
- Identification, distribution, and habitat of Coreopsis section Eublepharis (Asteraceae) and description of a new species.Journal of the Botanical Research Institute of Texas7(1):299‑310. https://doi.org/10.5962/bhl.title.63883
- Angiosperm Phylogeny Website. Version 12, July 2012 [and more or less continuously updated since]. http://www.mobot.org/MOBOT/research/APweb. Accessed on: 2016-8-15.
- Reasoning about taxonomies in first-order logic.Ecological Informatics2(3):195‑209. https://doi.org/10.1016/j.ecoinf.2007.07.005
- Towards best-effort merge of taxonomically organized data; pp. 151-154.2010 IEEE 26th International Conference of Data Engineering Workshops (ICDEW).2010 IEEE 26th International Conference on Data Engineering Workshops (ICDEW 2010),344pp. https://doi.org/10.1109/icdew.2010.5452756
- Merging taxonomies under RCC-5 algebraic articulations, pp. 47-54.Conference on Information and Knowledge Management. Proceeding of the 2nd International Workshop on Ontologies and Information systems for the Semantic Web (ONISW),Napa Valley, California.ACM,New York,118pp. https://doi.org/10.1145/1458484.1458492
- Merging sets of taxonomically organized data using concept mappings under uncertainty; pp. 1103-1120.Proceedings of the 8th International Conference on Ontologies, Databases, and the Applications of Semantics (ODBASE 2009). OTM 2009. Lecture Notes in Computer Science 5871. https://doi.org/10.1007/978-3-642-05151-7_26
- Reasoning About Taxonomies. Doctoral Dissertation.University of California at Davis,204pp. [ISBN978-1-124-22390-2].
- Taxonomy and irreproducible biological science.BioScience62(5):451‑452. https://doi.org/10.1525/bio.2012.62.5.3
- Keys to the flora of Florida – 10, Galactia (Leguminosae).Phytologia86(2):65‑74. URL: http://www.biodiversitylibrary.org/bibliography/12678
- A practioner's guide to concept mapping. Narrative for "Least Divisible Taxonomic Unit" approach to "concept mapping". http://sernec.appstate.edu/sernec-working-groups/how-map-taxonomic-concepts-alan-weakley. Accessed on: 2016-8-15.
- Applying concept mapping to 7,100 vascular plants. Presentation given at BIGCB Workshop, University of California at Berkeley, November 7-9, 2014. http://taxonbytes.org/wp-content/uploads/2014/10/Weakley-2014-BIGCB-Applying-Concept-Mapping-to-7100-Vascular-Plants.pdf. Accessed on: 2016-8-15.
- Flora of the Southern and Mid-Atlantic States.University of North Carolina Herbarium,Chapel Hill,1320pp. URL: http://www.herbarium.unc.edu/flora.htm
- New combinations, rank changes, and nomenclatural and taxonomic comments in the vascular flora of the southeastern United States.Journal of the Botanical Research Institute of Texas5(2):437‑455. URL: http://www.jstor.org/stable/41972288
- Darwin Core: an evolving community-developed biodiversity data standard.PLoS ONE7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
- Accountability and values in radically collaborative research.Studies in History and Philosophy of Science Part A46:16‑23. https://doi.org/10.1016/j.shpsa.2013.11.007
- Achievements and challenges in the integration, reuse and synthesis of vegetation plot data.Journal of Vegetation Science27:868‑879. https://doi.org/10.1111/jvs.12419
- Naming and contingency: the type method of biological taxonomy.Biology & Philosophy30(4):569‑586. https://doi.org/10.1007/s10539-014-9459-6
- Suppressing synonymy with a homonym: the emergence of the nomenclatural type concept in nineteenth century natural history.Journal of the History of Biology49(1):135‑189. https://doi.org/10.1007/s10739-015-9410-y
- Guide to the Vascular Plants of Florida, Third Edition.University Press of Florida,Gainesville,800pp. URL: http://upf.com/book.asp?id=wunde004# [ISBN978-0-8130-3543-7]
- Species level phylogeny of the genus Cornus (Cornaceae) based on molecular and morphological evidence - implications for taxonomy and Tertiary intercontinental migration.Taxon55(1):9‑30. https://doi.org/10.2307/25065525