|
Research Ideas and Outcomes : Project Report
|
|
Corresponding author: Sarah Faulwetter (sarifa@hcmr.gr)
Received: 08 Sep 2016 | Published: 12 Sep 2016
© 2016 Sarah Faulwetter, Evangelos Pafilis, Lucia Fanini, Nicolas Bailly, Donat Agosti, Christos Arvanitidis, Laura Boicenco, Terry Catapano, Simon Claus, Stefanie Dekeyzer, Teodor Georgiev, Aglaia Legaki, Dimitra Mavraki, Anastasis Oulas, Gabriella Papastefanou, Lyubomir Penev, Guido Sautter, Dmitry Schigel, Viktor Senderov, Adrian Teaca, Marilena Tsompanou.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Faulwetter S, Pafilis E, Fanini L, Bailly N, Agosti D, Arvanitidis C, Boicenco L, Catapano T, Claus S, Dekeyzer S, Georgiev T, Legaki A, Mavraki D, Oulas A, Papastefanou G, Penev L, Sautter G, Schigel D, Senderov V, Teaca A, Tsompanou M (2016) EMODnet Workshop on mechanisms and guidelines to mobilise historical data into biogeographic databases. Research Ideas and Outcomes 2: e10445. doi: 10.3897/rio.2.e10445
|

|
Abstract
The objective of Workpackage 4 of the European Marine Observation and Data network (EMODnet) is to fill spatial and temporal gaps in European marine species occurrence data availability by carrying out data archaeology and rescue activities. To this end, a workshop was organised in the Hellenic Center for Marine Research Crete (HCMR), Heraklion Crete, (8–9 June 2015) to assess possible mechanisms and guidelines to mobilise legacy biodiversity data. Workshop participants were data managers who actually implement data archaeology and rescue activities, as well as external experts in data mobilisation and data publication. In particular, current problems associated with manual extraction of occurrence data from legacy literature were reviewed, tools and mechanisms which could support a semi-automated process of data extraction were explored and the re-publication of the data, including incentives for data curators and scientists were reflected upon.
Keywords
biodiversity data, legacy literature, data archaeology, data rescue, text mining, biogeographic databases, data management
Introduction
Workshop "Tools, mechanisms and guidelines for the mobilisation of historical data into the systems"
To address problems associated with the extraction of species occurrence data from legacy biodiversity literature, EMODnet Biology Workpackage 4 (WP4) organised a workshop from 8–9 June 2015 in the Hellenic Centre for Marine Research in Heraklion, Greece. The aim of the workshop was threefold: a) to review the current problems associated with manual data extraction; b) to explore tools and mechanisms which could support a semi-automated process of data extraction and c) to discuss the re-publication of the data, including incentives for data curators and scientists.
Before the workshop, a list of old (ca. pre- 1930) faunistic reports, containing valuable occurrence data on marine species, had been compiled, and the data contained in several of these reports had been extracted manually by a team of data curators. During the data extraction process, the curators took notes on problems encountered and the time required to extract the data.
As data in legacy literature is presented in a variety of formats (tables, very verbose free-text, taxonomic sections) and varying levels of detail, the data curators presented an overview of the format of data and problems encountered during description, as well as the workflow required to transfer the data from a written report into modern digital formats.
The GoldenGATE-Imagine software was then demonstrated to the data managers participating in the workshop, followed by a short training session on how to semi‐automate the process of manual extraction of data. The software was tested on different types of legacy literature such as expedition reports, protocol logbooks and historical faunistic articles. GoldenGATE-Imagine was used both for digital born files and for scanned (image) PDF files.
The complete process from legacy literature identification to data publication via biogeographical databases was analysed via hands-on sessions: starting from how to scan a document, to import it into GoldenGATE-Imagine, to mark different document sections as well as entities of interests (e.g. taxonomic mentions and location names), to upload the markup to Plazi's TreatmentBank and from there to retrieve the auto‐generated Darwin Core Archives which in turn can be published through biogeographical databases.
Beyond hands‐on sessions, extensive discussions among the participants (bringing together data managers and information technology experts) resulted in the compilation of suggestions and best practices for data rescue and archaeology activities
The present report aims to summarise the outcomes of the workshop, but has also been enriched with conclusions and expertise acquired during subsequent digitisation activities carried out within EMODnet WP4. Specifically, the topics covered in this publication are:
- An overview of data archaeology and rescue activities carried out within the EMODnet consortium (section "LifeWatchGreece, EMODnet, and Lifewatch Belgium legacy literature data rescue") and the manual workflows currently being employed in these activities (section "Manual literature data extraction and digitisation workflow");
- A classification and evaluation of the problems encountered during the manual digitisation process (section "Common obstacles in manual occurrence data extraction"), and an estimation of the severity of these issues in a (future) software-assisted workflow (section "Potential problems in semi-automating the data extraction").
- A presentation of current tools, initiatives and approaches available to support the mobilisation of historical data (section "A software-assisted document annotation process and data publication")
- A evaluation of the GoldenGATE-Imagine software, after hands-on exercises by a group of data managers working on legacy data (section "EMODnet WP4 legacy document annotation using GoldenGATE-Imagine")
- A thorough discussion on possible improvements of the process of data mobilisation and downstream integration of data into literature and data repositories, including comments on current problems and recommendations for future practices.
Scientific background
Legacy biodiversity literature contains a tremendous amount of data that are of high value for many contemporary research directions (
Many of the above efforts have focused on extracting taxon names and parsing taxonomic (morphological) descriptions, called treatments. Treatments may include a variety of information on synonyms, specimens, dates and places, but in most cases follow a similar format, allowing algorithms to parse these blocks of information into sub-units (
In a time of global change and biodiversity loss, information on species occurrences over time is crucial for the calculation of ecological models and future predictions.Two major gobal biogeographic databases provide this information: the Global Biodiversity Information Facility (GBIF), which provides global index of biodiversity data, and the Ocean Biogeographic Information System (OBIS), focusing on marine biodiversity. While OBIS publishes the specific marine data, a global “common denominators” of such datasets (such as taxon, locality, date) also become available through GBIF as part of the global data pool.
This has also been recognised by the European infrastructure EMODnet (European Marine Observation and Data network). Within EMODnet Biology, Workpackage 4 (WP4) has been dedicated to data archaeology and rescue activities. The overall objective of WP4 is to fill the spatial and temporal gaps in marine species occurrence data in European waters. This is a two-part process of first identifying and locating data and then performing the steps required to digitise them and integrate them into a database, which can subsequently be distributed through publicly available data portals such as EurOBIS or the EMODnet data portal.
Extracting information from legacy literature: the manual procedure
LifeWatchGreece, EMODnet, and Lifewatch Belgium legacy literature data rescue
Legacy Literature Data Rescue activities are currently on-going in the framework of several research projects and were presented during the workshop:
- Within EMODnet WP4, four small grants were allocated for the digitisation and integration of selected datasets, contributing to a better coverage of underrepresented geographical, temporal or taxonomic areas (
Table ). These data were not available in scientific articles but as laboratory notes, field log books or other grey literature.1 - LifeWatch is the European e-Science Research Infrastructure for biodiversity and ecosystem research designed to provide advanced research and innovation capabilities on the complex biodiversity domain. Data rescue activities are ongoing in the framework of the LifeWatchGreece infrastructure (ESFRI) and are provided as an in-kind contribution to EMODnet WP4, and all rescued datasets are being propagated through the network of biogeographic databases, including the EMODnet Biology data portal. The activities of LifeWatchGreece focus on Greek and Mediterranean literature and target mainly historical publications (focus on the late 19th and early 20th century). These publications are mostly available digitally through the Biodiversity Heritage Library (BHL), but also scattered through various institutional and university libraries. Initially, an inventory of existing publications and datasets was compiled and prioritised for digitisation according to a number of criteria. Prioritised datasets were then described with extensive metadata, and finally the actual data were extracted and published. The following presents the progress status as of June 2015:
- > 220 historical publications / datasets identified
- ~70 of those chosen for digitisation
- > 50 annotated with metadata
- ~15 digitised and currently being quality-controlled and published
- The Flanders Marine Institute (VLIZ) performs ongoing data archeology activities in the framework of the Lifewatch Belgium project. These initiatives started in 2012, since then VLIZ has identified and recovered a number of historical biodiversity datasets, mainly from the Belgian part of the North Sea, but also datasets resulting from common Belgian-Kenyan research activities (
Table ).2
Datasets rescued under the EMODnet WP4 small-grant system
Title | Temporal coverage | Taxonomic coverage | Geographic coverage | Format of dataset |
Zooplankton Time series France - 1500 samples on a yearly basis | 1966 – present, yearly | Zooplankton | Western Mediterranean | Paper-based reports, grey literature |
Historical data on benthic macrofauna, demersal fish, and fish stomach content from the North Sea and the Baltic Sea | 1910-1952, yearly | benthic macrofauna | Limfjord, Denmark | Paper-based reports |
Romanian Black Sea Phytoplankton data from 1956 - 1960 | 1956-1960 | Phytoplankton | Black Sea | Paper-based report |
Romanian Black Sea Macrozoobenthos and Zooplankton and Recent Romanian Black Sea Macrozoobenthos | 1954-1968 and 1997-2014 | Macrozoobenthos, zooplankton | Black Sea | Paper-based datasets; non-standardised database |
Datasets rescued in the framework of Lifewatch Belgium (based on a slide by Simon Claus).
|
Biological datasets identified using the Belgian Marine Bibliography (2012)
|
Publication years: before 1995 Data extracted:
|
|
Biological datasets from Belgian-Kenyan research (2013)
| |
|
Phytoplankton data of the Belgian Part of the North Sea (2013–2014) Extraction focus: pigment & environmental variables, species observation data (plankton)
|
Publication years: 1968–1981 Data extracted:
Sources: Ijslandvaarten, Projekt Zee, Concerted Research Actions, Projekt Afvalwateren, Theses |
Manual literature data extraction and digitisation workflow
The process of manual data extraction follows a number of steps (
Workflow depicting the process of manually extracting data from legacy literature workflow, as currently performed in in EMODnet WP4. Abbreviations: OCR = Optical Character Recognition; OBIS = Ocean Biogeographic Information System; DwC = Darwin Core; IPT = Integrated Publishing Toolkit; medOBIS = Mediterranean Ocean Biogeographic Information System, GBIF = Global Biodiversity Information Facility
- Initially, candidate literature is identified, through library and literature research, and a copy of the publication is tracked down (either a hard copy or a digital version).
- The list of candidate literature is reviewed and prioritsed based on a list of criteria concerning the sufficency and adequacy of the information contained: taxonomic, spatial and temporal coverage and resolution, consistency of the information, presence/absence vs. abundance and presence of additional information (e.g. sampling methods). Another criterion is the language of the text. Historical publication are often written in a language other than English, and the data curator needs to be able to understand details on the data collection which often are presented in a verbose format. The document language might therefore limit the number of curators being able to process the data.
- If the data are in a paper-based format they are scanned (and sometimes OCRed - depending on the facilities of the holding library) to be accessible in a digital format.
- Extensive metadata are extracted for the selected document and registered by using an installation of the GBIF Integrated Publishing Tookit (IPT) repository. These metadata cover the title and abstract of the dataset, data collection methods, taxonomic, geographic and temporal coverage, associated persons, associated references and usage rights. The publication through the IPT ensures that, even if the data are not yet available, users are made aware of the existence of the publications, and they describe the data in enough detail to allow the user to judge whether it is worth obtaining the publication.
- The next step of the workflow is the manual data occurrence extraction from the document. The extracted pieces of information are transferred into a Darwin Core OBIS-compliant csv file which allows the information to be shared with other biogeographic information systems.
- During and after the extraction process, the data undergoes quality control. This includes the standardisation of taxon names (according to the World Register of Marine Species, WoRMS), cross-checking of coordinates, georeferencing of location, and data consistency checks, e.g. in terms of time, depth and abundance). Ideally, data are double-checked by another person.
- Finally, the data are published through the IPT installation along with their metadata. Data from the Mediterranean are published through the IPT installation of the Mediterranean Ocean Biogeographic Information System (MedOBIS), which is harvested regurlarly by EurOBIS and EMODnet and from where the data are subsequently integrated into OBIS and GBIF.
Common obstacles in manual occurrence data extraction
During the workshop an in-depth discussion, supported by examples, revolved around collecting feedback from the data curators detailing the difficulties they encountered during the data extraction process. The points which were presented and discussed are listed below:
- Distraction and errors: Data extraction is a slow and tedious process, curators reported a rate of approximately 3 pages per hour. While such work load can hardly be avoided, curators reported that distractions are the first cause of loss of time and producing errors: typographic errors, missing or mixed lines are frequent, and the necessary attention level cannot be kept high for a long time. As a consequence, productivity reduces as time passes. In addition, distraction and loss of concentration, as well as possible misinterpretation of information may cause errors. The data therefore need to be double-checked, ideally by another person. Therefore, the high demand in terms of time, concentration and workforce were identified as one of the major problems in digitising historical documents.
- Language: Many legacy publications are written in languages other than English, often limiting the availability of curators to process the text and data. The only solution to this problem is to assign, as far as possible, publications to curators that are able to understand the document language. A large team of multilingual curators is helpful in this aspect, but of course not always feasible.
- Availability of texts: Historical datasets are often difficult to locate, as they often exist only in print. Photocopies or scanned documents often are of of low quality (but see Biodiversity Heritage Library), not always OCRed, or the OCR text is of low quality. In these cases, information cannot be easily read (or, potentially, extracted by software) but needs to be rekeyed manually. Again, as many publications are only available via library loan or copies by the library, it depends on the digitisation facilities of the respective library to produce high-quality digital versions. During the workshop recommendations concerning requirements for digitisation and OCR were made (see below, section "Recommendations and Conclusions").
- Distribution of information across publications: Important information related to species occurrence (e.g. sampling station details, sampling methods) may exist in a different publication, which then needs to be located and (at least partly) processed, too. Expedition reports are often published in a series of volumes, and in those cases curators may need to complement the dataset by extracting information from two publications simultaneously.
- Distribution of information within a publication: Occurrence data can be either reported as free-text (taxonomic section), as a classification of taxa, in a table, or any combination of these (repetition of information), or part in text and part in table. The need to combine information contained in different manuscript elements and sections causes further delays.
- Inconsistency of information: The distribution or repetition of the same or complementary information across different sections of the same publication or even across publications often introduces inconsistencies and contradictory information on e.g. locations, dates and times, sampling methods, depths etc. Authors may also be inconsistent in the spelling and use of species names. Trying to resolve these contradictions causes significant delays in the data extraction, and is often not possible at all, as the deceased authors cannot be contacted any longer for clarifications.
- Unstructured information: Verbose and cumulative information is often encountered. As an example, all taxa of a certain family might be treated in one paragraph, but without structure (e.g. "Taxon A was found here, while taxon B was not found, it was found instead in place X"). Complex sentences and negations ("Argonauta was sought after, but not found") delay the extraction process and can result in errors if they are overlooked or misinterpreted. The debate about such negative data ("species X was not found") is still ongoing. There is relevant information (e.g. related to alien and invasive species) that can be derived from negative data, but it requires detailed information on the sampling methods, and there is no clear workflow to extract and represent such information from manuscripts and datasets.
- Mixed information: Legacy literature often reports both actual observation information and literature-derived occurrences. Often, this information is difficult to distinguish in free text reports, and data curators need to decide whether or not to include certain information in the final dataset.
-
Location information: Information on sampling locations can be either stations (with or without coordinates, either as a map or as a table), or named locations, or both (see
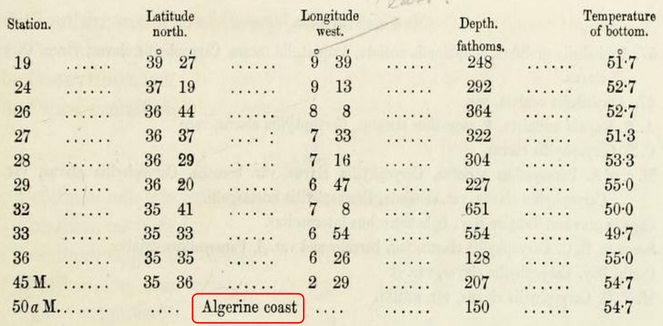
Fig. for an example). However, named locations may occur in different linguistic or historic versions (e.g. the historical "Candia" vs the contemporary "Crete" vs. the German "Kreta"). Thus, location information cannot always be checked against commonly used gazetteers, and to resolve these names and georeference them, additional, tedious and time consuming research into other literature is often needed. In other cases, coastlines or river courses may have changed over the decades or centuries, and given coordinates of a marine expedition may now fall on land and not in the sea. Such records have to be checked against old maps and the data have to be annotated so that they do not trigger errors in subsequent quality checks.2 -
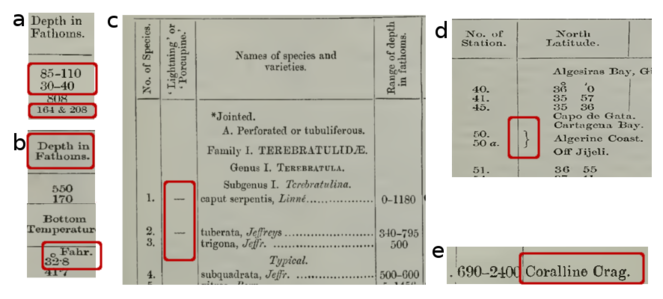
Depth measurements: For marine data, depth is an important feature, but depth measurements in historical literature must be treated with care. Often fathoms are given instead of metres (
Fig. a), sometimes without indicating the unit, or depth is expressed as verbatim info such as "shallow water". In other cases, sampling depths and/or bottom depths are reported without indicating what the depth measurement actually indicates. Depth ranges and more than one depth value are sometimes given for the same location or sample (3 Fig. a). To determine the actual bottom and/or sampling depths, bathymetric maps, information on sampling gear and unit conversions are required. While units can be converted, there is not always an agreement on the exact definition of descriptive terms such as “shallow”, “deep”, etc. Expressing the latter as a range or as standard vocabulary terms (e.g. using the Environment Ontology) is an alternative, but requires knowledge and a certain amount of informed interpretation by the data curators.3 -
Measurement units: Non-SI units are common in legacy texts. Depth can be expressed as fathoms, distances in miles or nautical miles, lengths as yards, feet or inches, temperatures in Fahrenheit (
Fig. a,b). Depending on the age and language (and thus cultural background) of the text, these units might not even correspond to those of the imperial system, but may be historic national or regional units of measurement. Verification of the used unit and its value and subsequent conversion into SI units can be time-consuming.3 - Format of species occurrences: Occurrence information might be either simple presence/absence or abundances (counts or densities) or biomass at a location, the latter sometimes split into sex and/or life stages. The format and typographical arrangement of this information is often difficult to understand, causing delays in data extraction. Often, no counts are given but estimates or arbitrary ranks of abundances such as "rare", "very rare", "common" "very common" and similar expressions. Such different types of information (presence/absence, counts, densities, measurements, estimates) require different representations in the final datasets.
-
Typography: Historical publications often use a variety of typographic arrangements, presumably to avoid repetition of data and/or save space (
Fig. ). Common symbols are ditto marks ("), indicating "same as above", two dots (..), indicating "no data", curly brackets (3 Fig. e) to unite two or more lines and hyphens and dashes (various meanings) (3 Fig. d). Abbreviations are common and are rarely explained. The typeface can be a challenge for OCR software, as symbols, letters and ligatures are often not represented correctly (3 Fig. c). Copying and pasting this information will introduce errors, so information has to be re-typed manually.3 - Mapping to data schemas: The information contained in (legacy) literature often describes complex relationships between stations, sampling events with possible replicates (and subsampling), gears and methods, various parameters, various depths and species, and commonly deviations from sampling protocols. These complex relationships are difficult to map to existing database or other electronic schemas (e.g. Darwin Core) and require experience in both the subject of the paper (e.g. sampling methods, taxonomy) and information managment, including databasing skills (see also paragraphs "Data encoding schema" and "Training of data managers" in the "Recommendations and Conclusions" section).
Stations without coordinates (red box) are commonly listed, as well as non-SI units, here: depth as fathoms (based on a slide by Aglaia Legaki, Gabriella Papastefanou and Marilena Tsompanou).
Examples of stylistic and typographic elements in legacy publications that delay the structured extraction of data: a) ranges or more than one value in one field; b) non-metric units which have to be converted to the SI system; c and d) unclear meaning of symbols; e) font type may cause problems in reading and/or optical character recognition (e.g. misinterpreting an “e” as “c” or “o”; "ll" as "11" or "U", "C" as "C" or "O") (based on a slide by Aglaia Legaki, Gabriella Papastefanou and Marilena Tsompanou).
Potential problems in semi-automating the data extraction
Prior to the workshop, a team of curators had assessed selected publications concerning their suitability for semi-automated data extraction. During this exercise, elements in the publications were identified which could potentially cause problems to a software attempting to automatically extract information. A briefing on the experience gained and example cases of issues encountered were presented to all participants and facilitated further discussion. Two basic categories of discussion topics were identified. The first relates to the quality of optical character recognition (OCR) and its application to reading legacy literature documents. The second refers to extracting occurrence information and to problems based on the authoring style, format and contents which may arise during semi-automated text extraction.
Optical Character Recognition
Historical publications are often not available in a "good" format, but either as photocopies or scanned documents of low quality. This prevents OCR software from correctly recognising certain characters in a scanned document (
Automated occurrence information extraction
Biodiversity legacy literature often contains complex natural language such as complex occurrence statements, negations, and references to background knowledge and to other expeditions, which can lead to false positive species-location associations and to incorrect occurrence extraction. Such ambiguity would still be present even in the case of 100% accurate digital text capture. Expert knowledge is often required to select the expedition-specific data occurrences and to interpret symbols, arrangement of information (e.g merged table cells, ditto marks, abbreviations).
A software-assisted document annotation process and data publication
To gain an overview of automated methods for species occurrence extraction and data publishing, the Plazi document annotation pipeline and the Biodiversity Data Journal (BDJ) were presented to the data curators by Donat Agosti and Lyubomir Penev, respectively.
Plazi rationale and taxonomic treatments
Plazi is an association supporting and promoting the digitization and publishing of persistently and openly accessible taxonomic literature and data. To this end, Plazi maintains literature and data repositories for taxonomic/biosystematic data, is actively involved in the creation of XML schemas and markup software to annotate and extract biodiversity information from literature, and develops new open access strategies for publishing and retrieving taxonomic information, including providing legal advice.
Taxonomic Treatments
"Nothing in biology makes sense except in the light of treatments" — Donat Agosti
A taxonomic treatment is a specific part of a publication that defines the particular usage of a scientific name by an authority at a given time. Typically, a taxonomic treatment can be seen as the scientific description of a taxon including a scientific name, often followed by e.g. references to older literature citing this taxon and putting it in relation to the current description (e.g. by defining synonymies, nomenclatural changes, etc). A treatment often contains a morphological description, citation of the studied materials (including references to the original specimen or observations used for the analysis) and additional information on the biology, ecology, host-relationships, etymology, geographic distribution, etc. of the taxon.
From a legal and information dissemination point of view, a taxonomic treatment is a discrete and coherent statement of facts constituting an observation of text extracted from the literature. Thus, it constitutes an observation and as such, the legal framework in many countries (e.g. USA, EU, Switzerland (
The Plazi document annotation pipeline
Plazi's aim of providing open access to marked-up taxonomic description and biodiversity information is supported by a pipeline of three components: a) the Biodiversity Literature Repository; b) the GoldenGATE-Imagine document editor and c) TreatmentBank, all described below:
The Biodiversity Literature Repository within Zenodo
Prior to making taxonomic treatments available to the community, the source document has to be included in the Biodiversity Literature Repository (BLR), which is a collection within the Zenodo repository (
Biodiversity related articles and instructions to the authors available on the Biodiversity Literature Repository home page.
GoldenGATE-Imagine
The GoldenGATE-Imagine (GGI) document editor is an environment for extracting, marking up, and annotating text and data from PDF files. GoldenGATE is a generic tool that can be highly customised. It can be used to convert single documents and customised to batch-process entire journal runs. The standard GoldenGATE editor allows importing text or html documents and is particularly suited for publications in the field of biological systematics, as it has enhancements for extracting and processing elements related to taxonomy and faunistics with an emphasis on taxonomic treatments. GoldenGATE-Imagine also reads (preferably born digital) PDF documents, performs OCR (and/or decodes embedded fonts), and then structures the document into smaller, hierarchical elements: pages, columns, blocks, paragraphs, lines, images, tables. A variety of (semi-)automated tools and manual markup and editing functionality are offered to further refine this identification of elements and semantic enhancement. The parser for document metadata (title, authors, journal, etc.) as well as bibliographic references can be customised to and find and tag those elements automatically. Bibliographic references are linked to the respective citation in the body of the publication. Simliarly, tables and figures are detected, analysed and made extractable. Their captions are linked to the respective table or image, and the figure and table citations to the respective captions.
GGI can detect and normalise taxonomic names and the higher taxonomic ranks added (backed by Catalog of Life (CoL), GBIF, and the International Plant Names Index (IPNI)), mark up the taxonomic treatments and their constituent parts, such as the nomenclature, description, discussion, distribution or etymology sections, and citations of other taxonomic treatments that can be annotated with the respective persistent identifier of the cited treatment. Named entities, such as collections codes, geo-coordinates, country names are routinely tagged (
The elements thus identified in a document are marked up with a generic XML used within TreatmentBank (see paragraph below) and stored as an IMF file (Image Markup File) which is a container that includes the source PDF, page images and a series of files that, together, show markup and annotations directly on the respective page image. Export functions allow for exporting the file as generic XML, as a Darwin Core Archive or to TreatmentBank.
TreatmentBank
TreatmentBank currently contains over 150,000 taxonomic treatments of ca. 17,000 articles. Articles from 18 journals are routinely mined, adding an approximate 100 treatments daily, resulting in an approximate 25% of the annually new described species. Depending on the degree of granularity, an array of dashboards is provided (
Each taxon treatment which is uploaded to TreatmentBank is assigned a persistent and dereferenceable http URI (e.g. http://treatment.plazi.org/id/3526F139-4833-C517-FF2F-0D406D6DF497) allowing users to cite the treatment. Individual treatments can be exported as XML, TaxonX schema based XML, (see paragraph below) or as RDF (Resource Description Framework). This allows users to share their markup work with the public. Access to treatments is open, whereas the original files and its various derived versions are only open to registered users. The data processing workflow, from a (legacy or prospective) publication to structured data is depicted in
Plazi workflow: from the publication through different levels of data processing to final availability of structured data.
Sharing data: TaxonX Schema, Darwin Core Archive and RDF
Treatments in TreatmentBank can be accessed in different formats. TaxonX (
Data papers and the Biodiversity Data Journal
Data papers are “scholarly publication of a searchable metadata document describing a particular on-line accessible dataset, or a group of datasets, published in accordance to the standard academic practices” (
Given the the previous two definitions, a data paper could complement a legacy-literature-extracted species occurrence dataset release in an ad hoc repository such as GBIF and OBIS, increase outreach and facilitated retrievability (see also section "Data publication landscape" below).
The Biodiversity Data Journal (BDJ) builds around such data paper concept and aims to provide both a workflow and an infrastructure. Through the act of scholarly publishing, data are mobilised, peer-reviewed, standardised (and thus made interoperable) and widely disseminated. All structural elements of the articles —text, morphological descriptions, occurrences, data tables, etc.— are marked up and treated and stored as data (see also Data Publishing Policies and Guidelines of Pensoft Publishers (
Re-publication of historic datasets in a modern, standardised digitised form is encouraged by journals such as the BDJ, and pecularities of such publications (e.g. authorship) were discussed during the workshop. Overall, participants agreed that the re-publication of digitised legacy data as data papers could provide an incentive for curators and scientists to get involved into digitisation activities (see also section below "“Reward” of data curators").
The latest development towards providing sustainability of publications in BDJ is its integration with Zenodo. Currently, all articles published in BDJ (and all other Pensoft journals) are automatically deposited in the Biodiversity Literature Repository collection in Zenodo upon publication.
EMODnet WP4 legacy document annotation using GoldenGATE-Imagine
After the presentation of the GoldenGATE-Imagine editor, participants had the opportunity to work with the software and evaluate it regarding its suitability for data extraction from legacy literature. The tutorial followed in this workshop was based on the GoldenGATE-Imagine Manual.
Five historical publications, all available through the Biodiversity Heritage Library, were used to test the software:
-
Calman (1906) -
Duncan (1873) -
Jeffreys (1882) -
Laackmann (1913) -
McIntosh (1876)
These publications had been scanned by the Biodiversity Heritage library and are available in a variety of formats (image, text, PDF). In addition, a digital-born publication was used for demonstration and training purposes. Participants learned to automatically segment a text into pages, blocks, columns, treatments, images and tables, to extract metadata and references and to markup taxonomic treatments and the information contained within (in particular occurrence information). The marked-up information was then extracted as a DarwinCore Archive.
Evaluation of the semi-automated annotation process
After the training session, participants provided feedback on the use of GoldenGATE-Imagine and its usefulness for the purposes of mobilising data from legacy publications. General remarks, both from data curators and other particpants were:
- Optical Character Recognition is a problem with PDF files retrieved from BHL. Loading and processing of these files in GGI was time-consuming and error-prone.
- A possible improvement of GGI could be its adaptation to open e.g. a .zip file containing image files instead of PDFs, which result from scanning.
- The OCR effort could be pushed from 5 down to ca. 2 minutes per page with experience/GGI improvements.
- Marking up documents has a slow learning curve and is different for each new document with a different structure of the information. The longer the document, the faster the progress.
- The data table extraction was considered a very useful tool of GGI.
- GGI is customisable by both developers and users with a little technical know-how. Thus, an occurrence-extraction specific version of GGI could be spinned out.
- Around 48% of the taxonomic names found in documents processed by Plazi are not known to GBIF. This implies a great potential for new contributions of taxonomic names to the global registers by initiatives such as data rescue from legacy literature.
In addition to the informal discussions, GoldenGATE-Imagine was also formally evaluated. A questionnaire was handed out to the users after the training session (questionnaire proposed by the BioCreative IV Interactive Annotation Task to evaluate system usability (
Given the low sample size (N = 8 complete questionnaires returned), of which only one was by an experienced user, results are here only presented in a descriptive way (
Results of the evaluation questionnaire submitted to the participants of the workshop after a demonstration of GoldenGATE-Imagine software; see text for explanation of how scores are calculated.
Group of questions | Potential range | Median | Score |
G1. Overall reaction | 40 – 120 | 80 | 94 |
G2. Overall comparison with similar systems | NA | NA | NA |
G3. System's ability to help complete tasks | 12 – 60 | 36 | 45 |
G4. Design of application | 32 – 160 | 96 | 108 |
G5. Learning to use the application | 24 – 120 | 72 | 67 |
G6. Usability | 40 – 200 | 120 | 125 |
Evaluations of the questionnaire were provided on a Likert scale (1-5), with no need of reversion (as all questions were formulated with a positive statement). The overall score is presented per group of questions (G1– G6) along with its potential range: the minimum of the range is defined by the value 1 assigned to all questions and the value 5 assigned to all questions, multiplied by the number of responses gathered (varied per question) and summed for each group of questions. The median of these ranges was then compared with the score obtained in the evaluation questionnaire.
While a positive (above the median) and negative (below the median) score are clearly expressing a positive and a negative trend respectively, an average score could a) result from two distinct, contrasting groups of opinions (e.g. half of the participants scored 1 and the other half scored 5 the same question) or b) indicate a true neutra lity. In our case, scores were concordant among participants: the slightly positive/positive evaluation of G1; G3; G4 (above the median) resulted from values ranging from 3–5 assigned to the single questions, while a majority of "3" values defined the neutral opinion obtained for G5 and G6.
Combining the results of the questionnaire with the feedback provided during the discussions in the workshop, participants saw potential in using the software for supporting data extraction activities, however, the learning process is initially slow, and not all documents seem equally suitable for software processing.
Recommendations and conclusions
The following conclusions and recommendations emerged from the discussions throughout the meeting, and from experiences gathered throughout the activities of EMODnet WP4. By taking note of all the obstacles towards digitisation and possible solutions to overcome them, coming from good practices, we hope to provide insights for further developments and more efficient work. Issues are presented along with respective solutions/mitigations proposed by the participants.
OCR best practices and BHL scanned documents
Problems with OCR in old documents are very common. In some cases it may be more efficient to manually rekey the original text rather than to edit the scanned image. If the document is not already digitised, it is recommended to create a scan of the highest possible quality. Outsourcing of the document scanning to a specialised company is suggested, especially if larger volumes of literature are to be scanned. Plazi is investigating contracting with commercial companies to perform text capture and of historical publications and providing digital versions encoded using the Journal Article Tag Suite (JATS) to use as input into GoldenGATE for application of domain specific markup. For in-house document scanning some practical tips are listed below. Nevertheless, it is well worth getting professional input, as scanning and digital text capture should not be a task for data curators and/or biologists. Even if documents have different characteristics which make generalisations difficult, a few general guidelines for scanning can be derived from successful experiences:
- For older book pages (19th and 20th century) capturing in color and OCRing gives more accurate results than grayscale or bitonal. The files can always be converted to bitonal after OCR (if necessary for storage limitations).
- For book digitisation images should be captured at a minimum of 400 ppi (at 100% item size). If the font size is particularly small or complicated, images should be captured at 600 ppi (but 400 ppi is the recommended minimum – experience by The Digital Imaging Lab).
- If a 35 mm camera is available (16, 24 or 36 megapixels), the frame should be filled as much as possible and then downsampled to 400 ppi. This usually give a sharper and more detailed image than capturing the objects original size at 400 ppi (Dave Ortiz, pers. comm.). However, the use of macro- or copy-lenses is required to prevent distortion of the text at the edges (“rounded squares”).
- Non-necessary parts of the document can be omitted for the sake of relevant ones: spending an initial amount of time for evaluating the document and locating the points of interest can save time later and allow the data manager to work on high quality scans.
In summary, suggested specifications for scanning are listed in
Recommended OCR book scanning specifications.
| Scanning mode | RGB color |
| Scanning Resolution | 400 ppi (at 100% of object's size) |
| Output format | TIFF |
| Color Depth | 48 bit |
In case an already scanned document needs to be retrieved from BHL, it is recommended that the corresponding document is retrieved from the Internet Archive as a JP2 (jpeg2000) version (
Top: to retrieve a scanned BHL book document from BHL click on the “Download Contents” icon on the top-right and select to browse the corresponding web page on the Internet Archive (“View at Internet Archive”). Bottom: The link to the jpeg2000 (JP2) image is found on the bottom right. Sources: top: http://biodiversitylibrary.org/page/9663476; bottom: https://archive.org/details/mittheilungenaus17staz.
The scanning process itself appears to be a bottleneck in terms of the quality, size, resolution etc. of a scan. These factors are, however, crucial for the quality of the OCR process. Expert knowledge on “scanning best practice” should be obtained; this is also important for the usefulness of GoldenGATE-Imagine, as otherwise users might experience frustration. Due to these constraints not all documents are suitable for semi-automated processing: some documents are simply too complex to be processed by software. A recommendation for best practice is therefore is to seek advice at the starting phase, to classify documents according to a scale of simple to complex, and from do-able to impossible, and then set up a workflow that will allow massive and fast assisted data extraction.
“Reward” of data curators
Having to deal with a huge amount of work and constraints affecting the speed and efficiency, data curators should be given incentives to pursue their data rescue efforts. Publishing the outcomes of their work and being cited when the extracted data are used in other analyses is one of the most obvious incentives. Re-publishing data of historical publications allows these papers to be shareable and searchable, offering baselines for current research. Credit should, therefore, be given to people who made these valuable data accessible again, i.e. the data curators.
A high-quality publication of the digitisation efforts would need to comprise a description of the legacy documents, the rescue / digitisation methodology, and the actual data extraction and quality control process along with the results (the actual data). In addition to publishing the species occurrence data through GBIF/OBIS, linking the results to Plazi taxonomic treatments could add value and strengthen the outreach of the extracted datasets. The publication as a data paper (e.g. in BDJ) could be assisted by an integrated workflow, e.g. from annotation in GGI to publishing in BDJ. Emphasis should not only be given to the initial publication of a dataset, but also to the ability to incrementally include annotations, corrections, and additional elements (e.g. tables, maps) once these have been established.
Data publication landscape
Data papers are strongly recommended given the emerging success of open data (
Open Data: an emerging landscape of data and other academic publications (based on a slide by Dmitry Schigel).
However, peer-review of data papers poses some new challenges: not only needs the actual text of the publication to be reviewed, but also the data themselves. Towards this end, expertise from different fields is required: a biologist, ecologist or an oceanographer needs to assess the usefulness of the data for potential models and analysis, for datasets including taxonomic information a taxonomic expert may be required. To evaluate the quality of the data, it is moreover advisable to include a reviewer familiar with data digitisation and/ or quality control procedures. These procedures will need to be addressed and streamlined in the future, and Plazi and BDJ are committed to developing tools and pipelines that could facilitate the process.
Finally, the utmost criterion for the quality of the data is their use after they are published. For this reason a "data impact factor" system should be established and implemented, based on the views, downloads and use cases of the published data. To initate the discussion, it is considered that the current landscape of factors such as the impact factor, h-index and citation index, provides a suitable basis for such a discussion to start.
Data encoding schema
A lesson learned from the manual digitisation was the inadequacy of the Darwin Core format for encoding and digitising. Most data curators involved in the digitisation activities had received only basic training before the activities (see paragraph below, “Training data managers”), and the Darwin Core schema had been proposed to them for ease of use. However, Darwin Core is a data exchange format that theoretically should be generated and read only by computers; only the matching phase should be performed by a human. This schema forces data managers to repeat the same information sometimes in thousands of rows. During quality control of the data —despite all the care that data managers had taken—, many inconsistencies were discovered in these repeated lines, especially in long text fields (e.g. reference citation). A highly common mistake is the auto-completion of cells by the spreadsheet software, generating a +1 increase if the last character in the cell is a digit (e.g. for authorship, the consecutive rows for the same scientific name is Linnaeus, 1758 / Linnaeus, 1759 / Linnaeus, 1760, etc.). Thus, certain types of checks have to be performed systematically for all records for all text fields, which significantly lengthens the quality control procedure.
In addition, the data structure extracted from a paper is a subset of a very complete and complex schema of sampling events taking into account various gears, various parameters, various depths with possible replicates (and subsampling). Unless they are very experienced, data managers have difficulties to fit these complex interactions of stations, sampling and replicate codes into a database or other electronic schema (e.g. DwC), as each paper has its own peculiarities.
Therefore, it is recommended to assist less experienced data curators at the start of the data encoding process by establishing establish a schema that minimises the repetition of identical data and reflects as closely as possible the structure of data in papers. Then, the integration into a final database (e.g. MedOBIS, EurOBIS) should be done by a (team of) professional data manager(s), who also perform the final —and minimal— quality control. To share the data with other repositories, Darwin Core Archives can be generated automatically, e.g. through an IPT (GBIF Internet Publishing Toolkit) installation.
Training data managers
Training data managers is very challenging (and costly), especially when trainees are not accustomed to a databasing mindset. To fulfill the obligations of data management activities in LifeWatchGreece and EMODnet Biology WP4, about 25 data managers had received basic training, but it is not expected that more that 20% of them will continue any data digitisation activities after the end of the project. Thus, training should be kept at a minimum level and supported by tools and infrastructures, as outlined above (paragraph “Data encoding schema”), and intensive training should rather target data managers who will continue to encode data long after the end of the project or training.
A plea for professional data manager position in research institutes
From the recommendations about the data schema and the training, there is one logical conclusion: the number of professional, permanent data manager positions in academic institutions need to be increased. Training data managers during 3-years projects is not efficient in the long-term regarding data encoding speed and data quality. In particular, quality control requires much experience to be thorough and reach an operational high level. Large repositories such as GBIF, OBIS, FishBase, and others are often criticised to deliver data of a low quality level (e.g.
In the era of Big Data in the biodiversity domain, and if the targeted goals are to aggregate, share and publish as many of good quality data as possible, each biodiversity research institute should have one or several professional data managers, helping researchers and technicians to create good quality datasets, well curated and documented, to be subsequently published through large global databases such as OBIS or GBIF. This has been proven by the success of WoRMS and FishBase cases among others, where some data managers are employed for more than ten and 25 years respectively, and is a practice which should be adopted by the scientific community at large.
Final conclusions
Overall, the high importance of data locked up in legacy biodiversity literature was acknowleged by all participants. Currently, extracting this data to make it available through global biogeographic databases, is a manual, tedious, costly and error-prone process. However, tools are available that could assist in mobilising this data: high-quality scanners to produce digital versions of historical publications, document editors to identify and extract the required information, and publishing platforms that help to integrate and disseminate the data to the wider public. Currently, none of these tools is tailored to the processing of legacy literature and data archaeology, and bottlenecks and difficulties still exist that prevent the massive semi-automated extraction of historical data. Future research efforts therefore need to go into adapting and fine-tuning the existing tools and integrating them into a pipeline that allows for a smooth workflow: from locating valuable historical publication to scanning, data extraction and quality control and finally the publication of an integrated report of both the rescue activities and the resulting dataset. To reach this goal, expertise is required from a broad range of domains: from librarians to imaging experts, from biologists to data managers, computer scientists and finally experts on data publishing and integration.
Funding program
This paper was supported by the European Marine Observation Data Network (EMODnet) Biology project, funded by the European Commission’s Directorate—General for Maritime Affairs and Fisheries (DG MARE). Additional financial support was provided by the LifeWatchGreece infrastructure (MIS 384676), funded by the Greek Government under the General Secretariat of Research and Technology (GSRT), National Strategic Reference Framework (NSRF).
Grant title
The European Marine Observation and Data Network (EMODnet) is a long-term marine data initiative of the European Union. It comprises seven broad discipliniary themes: bathymetry, geology, physics, chemistry, biology, seafloor habitats and human activities. The aim of the initiative is to assemble, harmonise, standardise and quality control marine data, data products and metadata within these thematic areas and to integrate the fragemented information into a central portal, through which the information is freely available.
The LifeWatchGreece Research Infrastructure is a comprehensive data and analysis infrastructure providing access to biodiversity and ecology data of Greece and South-East Europe. An integrated platform offers both electronic services (e-Services) and virtual labs (vLabs) to facilitate access to data and analysis tools. These allow large scale science to be carried out at all possible levels of the biological organisation, from molecules to ecosystems.
Hosting institution
The workshop was hosted at the Hellenic Centre for Marine Research in Crete, Greece.
Author contributions
This publication is based on the workshop minutes which were compiled on-the-fly during the workshop by all participants in a shared online document. The minutes were compiled into a final workshop report by Evangelos Pafilis, Lucia Fanini, Nicolas Bailly and Sarah Faulwetter. All other authors contributed by providing presentations, discussions and input during the workshop and afterwards during the compilation of this publication, and are listed in alphabetical order.
References
-
Taxonomic information exchange and copyright: the Plazi approach.BMC Research Notes2(1):53. DOI: 10.1186/1756-0500-2-53
-
Making Data a First Class Scientific Output: Data Citation and Publication by NERC’s Environmental Data Centres.International Journal of Digital Curation7(1):107‑113. DOI: 10.2218/ijdc.v7i1.218
-
The Cumacea of the Puritan Expedition.Mittheilungen aus der Zoologischen Station zu Neapel17:411‑432. DOI: 10.5281/ZENODO.14941
-
TaxPub: An Extension of the NLM/NCBI Journal Publishing DTD for Taxonomic Descriptions. In: Bethesda (MD): National Center for Biotechnology Information (US); 2010Proceedings of the Journal Article Tag Suite Conference 2010. URL: https://www.ncbi.nlm.nih.gov/books/NBK47081/
-
The data paper: a mechanism to incentivize data publishing in biodiversity science.BMC Bioinformatics12:S2. DOI: 10.1186/1471-2105-12-s15-s2
-
A Description of the Madreporaria dredged up during the Expeditions of H.M.S. Porcupine‘ in 1869 and 1870.The Transactions of the Zoological Society of London8(5):303‑344. DOI: 10.1111/j.1096-3642.1873.tb00560.x
-
Using legacy botanical literature as a source of phytogeographical data.Plant Ecology and Evolution148(2):256‑266. DOI: 10.5091/plecevo.2015.1048
-
Detailed mark-up of semi-monographic legacy taxonomic works using FlorML.Taxon63(2):377‑393. DOI: 10.12705/632.11
-
On the Mollusca procured during the ‘ Lightning’ and ‘Porcupine’ Expeditions, 1868-70.Proceedings of the Zoological Society of London50(4):656‑688. DOI: 10.1111/j.1096-3642.1883.tb02779.x
-
Adriatische Tintinnodeen.Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften. Mathematisch-Naturwissenschaftliche Klasse122:123‑163. URL: http://biodiversitylibrary.org/page/36050965
-
Digitising legacy zoological taxonomic literature: Processes, products and using the output.ZooKeys550:189‑206. DOI: 10.3897/zookeys.550.9702
-
Data patterns in multiple botanical descriptions: Implications for automatic processing of legacy data.Systematics and Biodiversity1(2):151‑157. DOI: 10.1017/s1477200003001129
-
BioCreative IV Interactive Task.Proceedings of the Fourth BioCreative Challenge Evaluation Workshop.,1.190-203pp. URL: http://www.biocreative.org/media/store/files/2013/bc4_v1_27.pdf
-
On the Annelida of the Porcupine Expeditions of 1869-1870.Transactions of the Zoological Society of London9:395‑416. DOI: 10.1111/j.1096-3642.1976.tb00244.x
-
Integrating and visualizing primary data from prospective and legacy taxonomic literature.Biodiversity Data Journal3:e5063. DOI: 10.3897/bdj.3.e5063
-
Evolutionary informatics: unifying knowledge about the diversity of life.Trends in Ecology and Evolution27:94‑103. DOI: 10.1016/j.tree.2011.11.001
-
Pensoft Data Publishing Policies and Guidelines for Biodiversity Data. http://community.gbif.org/mod/file/download.php?file_guid=13043
-
XML schemas and mark-up practices of taxonomic literature.ZooKeys150:89‑116. DOI: 10.3897/zookeys.150.2213
-
Global biogeographical data bases on marine fishes: caveat emptor.Diversity and Distributions14(6):891‑892. DOI: 10.1111/j.1472-4642.2008.00519.x
-
Semi-automated XML markup of biosystematic legacy literature with the GoldenGATE editor.Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing12:391‑402.
-
Data issues in the life sciences.ZooKeys150:15‑51. DOI: 10.3897/zookeys.150.1766
-
Applications of Natural Language Processing in Biodiversity Science.Advances in Bioinformatics2012:1‑17. DOI: 10.1155/2012/391574
-
From XML to XML: The why and how of making the biodiversity literature accessible to researcher.Language Resources and Evaluation Conference (LREC)May 2010, Malta:1237‑1244. URL: http://oro.open.ac.uk/20856/1/787_Paper.pdf