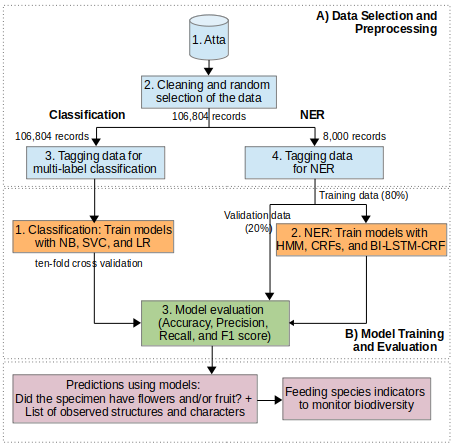
The proposed general workflow includes two phases: A) Data Selection and Preprocessing using the Atta database (INBio). First, the data were cleaned by removing duplicate records, records written in English and null morphological descriptions, amongst other processes. Then, two datasets were selected for the next phase, one for Classification and one for NER. Those datasets were used for training and test activities. B) During the Models Training and Test phase, models were generated using algorithms such as: Multinomial Naive Bayes (NB), Linear Support Vector Classification (SVC) and Logistic Regression (LR) for Classification and Hidden Markov Model (HMM), Conditional Random Fields (CRF), and Bidirectional Long Short Term Memory Networks with CRF (BI-LSTM-CRF) for NER. Metrics like accuracy, precision, recall, and F1 score were used to test them.