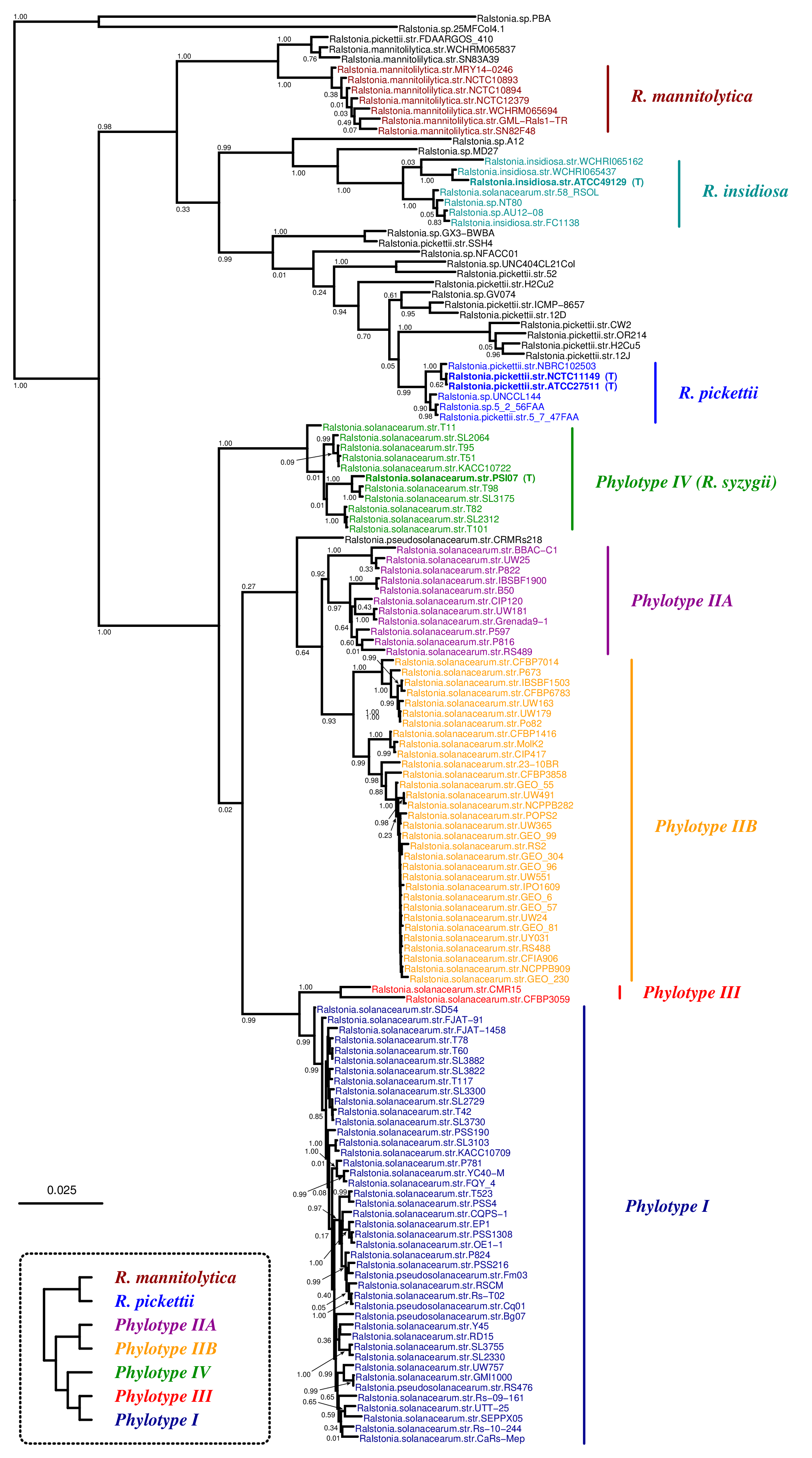
Phylogenetic tree inferred by JolyTree from the RefSeq genomes belonging to the genus Ralstonia. For each type strain (in bold), the clade determined by the isolates expected to belong to the same species (e.g. estimated pairwise distances < 0.05) is labeled by the species name and colored accordingly. Leaf names were automatically generated. Scale bar corresponds to an estimated evolutionary distance of 0.025. The inset summarizes a model tree of the Ralstonia species based on the phylogenetic analysis of