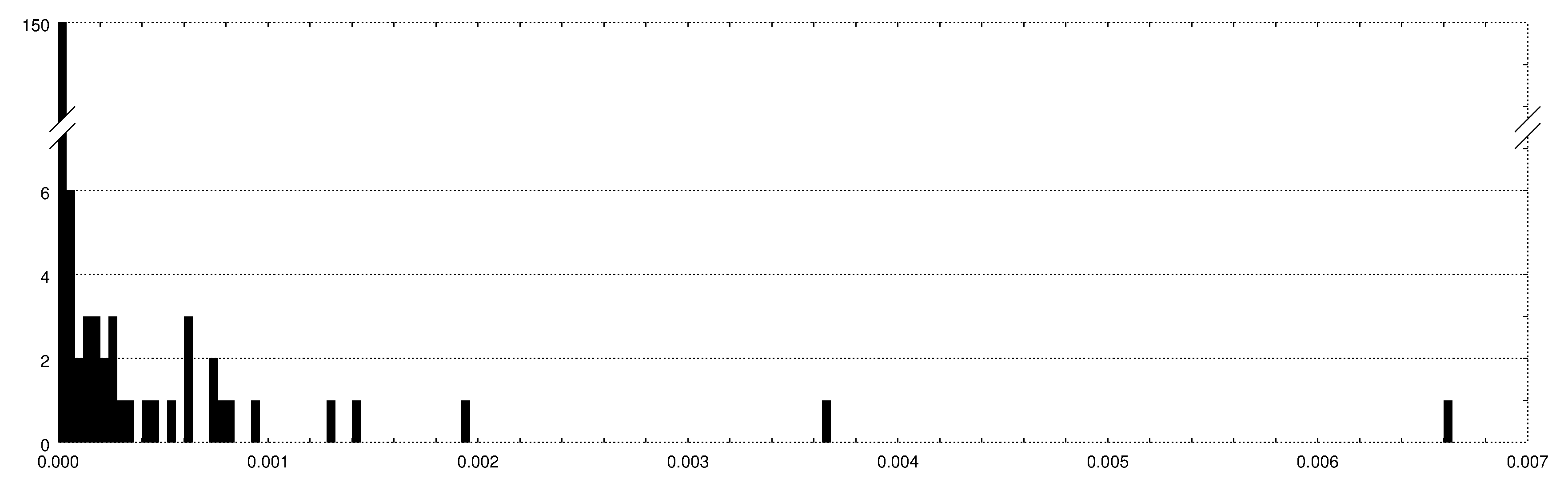
Distribution of the normalized Balanced Minimum Evolution (BME) tree length differences observed between phylogenetic trees inferred by FastME with and without the data noising strategy implemented by JolyTree. This distribution represents 187 observed values \(\Delta(T_0, T) = (\ell(T_0)-\ell(T))/\ell(T_0)\), where \(T_0\) is the tree inferred by FastME alone, \(T\) the one inferred by JolyTree via the data noising strategy, and \(\ell(T)\) the BME tree length estimate of \(T\). A tree \(T\) is considered as more accurate that \(T_0\) when \(\Delta(T_0, T)\) > 0.