|
Research Ideas and Outcomes : Research Idea
|
|
Corresponding author: João Vitor Dutra Molino (molino@usp.br)
Received: 25 May 2016 | Published: 23 Jun 2016
© 2016 João Vitor Dutra Molino, Tiago Lubiana Alves, Livia S Ferreira-Camargo, Miguel A Croce, Allan Tanaka, Felipe X Buson, Pedro de Freitas Ribeiro, Antony B Campos-Salazar, Eduardo Padilha Antonio, André Schraider Maizel, Viviane M Siratuti, Claudia Costa, Samarina R Wlodarczyk, Raquel de Souza Lima, Fabio N Mello, Stephen P Mayfield, João C M Carvalho.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Molino J, Lubiana Alves T, Ferreira-Camargo L, Croce M, Tanaka A, Buson F, Ribeiro P, Campos-Salazar A, Antonio E, Maizel A, Siratuti V, Costa C, Wlodarczyk S, de Souza Lima R, Mello F, Mayfield S, Carvalho J (2016) Chimeric spider silk production in microalgae: a modular bionanomaterial. Research Ideas and Outcomes 2: e9342. doi: 10.3897/rio.2.e9342
|

|
Abstract
Background
In this project, we propose to explore the modular characteristic of spider silk proteins, through synthetic biology techniques, by combining and directing its properties to the desired application. The aim of this project is to generate a modular bionanomaterial able to immobilize proteins. This bionanomaterial will be composed of modular recombinant proteins from spider silk, which will be the immobilization support to other proteins, in this project an antimicrobial protein (enzybiotic). By combining these proteins and their properties, the primary focus will be the use of this technology for the development of artificial skin for burn victims.
New information
The recombinant proteins, spider silk proteins and enzybiotics, will be expressed in Chlamydomonas reinhardtii strains by nuclear transformation. Each recombinant strain will express a different protein, which will contain the N- and C-terminal polymerization domains from native spider silk proteins. These domains are essential to the polymerization step and, subsequently, for production of a material very similar to silk. This material will be evaluated regarding its antimicrobial and mechanical properties, as well as the system productivity. These results may shed some light on spider silk-based immobilization support effectiveness, even for other biotechnological applications, such as the one idealized here.
Keywords
Bionanomaterial, Silk protein, Recombinant protein, Microalgae, Enzybiotic
Overview and background
Immobilization techniques are applied to a wide range of treatments and processes, from medical applications to biotransformations in industrial plants. The industrial/commercial application of biomolecules such as proteins depends on the stability and functionality of the process employed. This process often differs from the natural environment of proteins in terms such as temperature, presence of organic solvents and pH values. Consequently, techniques such as immobilization can promote stabilization and add functionality even when these biomolecules are under different environmental conditions. This stabilization is normally achieved by protein binding to a scaffold (
The aim of this project is to use this biomaterial for protein immobilization. Initially, we will immobilize enzybiotics for application to burn wounds, as a model to test this support. However, there are other possible applications with economic and academic interest.
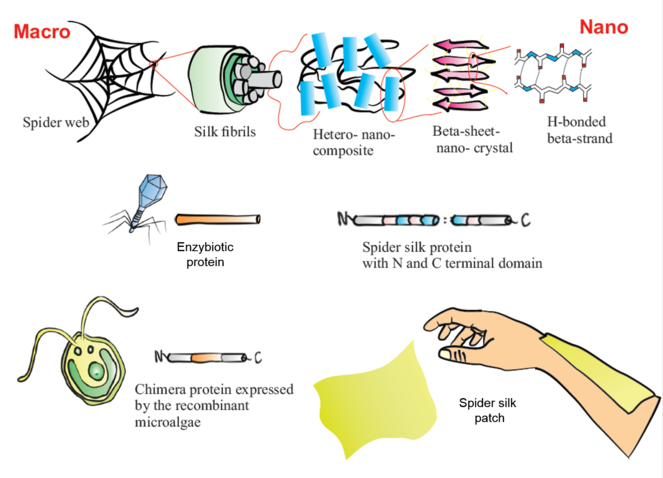
Project overview. Schematic representation of spider web structure from macro to nano scale. A representation of: enzybiotic protein from a bacteriophage; a spider silk protein with repetitive domains and N and C terminals; host expression system Chlamydomonas reinhardtii and a chimeric protein envisioned in this project; and the final product, a biopatch produced from recombinant silk proteins and chimeric proteins.
Immobilization Systems
The retention of molecules inside a reactor, or an analytical system, is described as immobilization, whose purpose is to improve protein stability, selectivity, and particularly for the enzymes, increase catalytic activity (
Enzymes are widely used in our society, and their applications range from industrial processes - such as in food production, biofuels and tissue - to more complex therapeutic applications, such as biopharmaceuticals (e.g., asparaginase use for acute lymphoblastic leukemia treatment (
A medical application that benefits from enzyme immobilization is biosensor development (
Enzybiotics
Enzybiotics comprise a class of enzymes with antibiotic activity. These enzymes are able to fight resistant bacteria, such as MRSA, VRSA and VISA (
Spider webs
The polymer constituting the spider silk bears interesting properties for various applications, including immobilization of molecules such as proteins. Spider silk is known mainly for its tensile strength and fracture resistance, but also exhibits elasticity, adhesion, biocompatibility and low degradation. Its strength can be compared to Kevlar synthetic polymer, which is composed of aramid and is used in for manufacturing body armor (
It is known that certain repetitive sequences of amino acids confer specific properties to these structures and proteins in tissue, allowing one to obtain materials with desired characteristics through genetic manipulation of these structural domains. The poly-alanine domains (poly(A/GA) (Glycine-Alanine) in MaSp1 proteins, MaSp2 and MISP are associated with formation of beta-sheets and the production of strong fibers, while repeating sequences "GPGGx" and "GGX" as in Flag protein, preferably generates an elastic beta-spiral region, which provides elasticity (
In addition, terminal domains (N-terminal NT and C-terminal CT) are highly conserved both among species and different types of silk (
There is some evidence of dependence on N- and C-terminal domains for polymerization, which can lead to interesting technological possibilities. The adaptation of these domains flanking a protein of interest opens the possibility of its immobilization if spun alongside “native” spider silk proteins. Moreover, core structural domains’ ("poly(A/GA" and "GPGGx" and "GGX") customization influence the physical properties of the silk (
Expression System
Heterologous proteins are produced in several established expression systems, such as Escherichia coli, mammalian cell lines and yeast, but several systems are in the developing pipeline. Appropriate expression system choice should be based on priorities regarding performance and requirements of each recombinant protein (
Therapeutic proteins are preferably produced in transgenic mammalian cell systems, because of their ability to express and correctly fold proteins. However, its production cost is high, especially when compared to plants as expression systems. Molecules such as monoclonal antibodies (mAbs) are mainly produced in mammalian cells and their average production cost in this system is estimated to be $ 150.00 per gram of raw materials, whereas production in plant systems costs approximately US$0.05 per gram (
However, the studies on genetic engineering using microalgae are incipient and present challenges. The main challenge is the low productivity of recombinant proteins expressed in the nuclear genome, hindering commercial applications to date (
Objectives
- Evaluate production capacity of synthetic spider silk proteins (based on MaSp1 and MaSp2) and protein chimeras with NT and CT domains flanking enzybiotics in Chlamydomonas reinhardtii by transforming the nuclear genome;
- Set methods for polymerization of silk produced by microalgae;
- Test biopolymer and antibiotic properties of spun spider silk, pure and in combination with enzybiotic chimeric proteins;
- Assess the project development in an iGEM competition context regarding its scientific achievement and the real-time openness in all process steps: idealization, laboratory procedures, results and discussion
Impact
Many obstacles need to be overcome for the effective production of a biomaterial such as a recombinant spider silk capable of immobilizing proteins in its matrix. With this purpose, this project offers solutions, as of yet untested, for example: the use of microalgae as a production platform for the expression of spider silk proteins, as well as chimeras with NT and CT domains, and flanking enzybiotics. Research on transgenic microalgae are driven by the global demand for recombinant proteins and other bioproducts. This biotechnological market is growing exponentially, it has reached 140 billion dollars in sales as of 2013, and it continues to grow (
More than developing a product that could help thousands of patients, the development of an antibiotic chimeric biopolymer in C. reinhardtii can result in many scientific outcomes. Spider silk is a challenging material to work with, and accessing its production with chimeric and functional materials can pave the way for many applications in the biomaterial field. Moreover, the microalgae community is still incipient, in comparison to the E. coli system, and integrative projects like this one can expand the synthetic biology tools applied to this model microorganism, which will benefit the scientific community.
It is important to highlight the fact that this project will be carried out at an international competition dedicated to the development of high-level research in Synthetic Biology (iGEM - International Genetically Engineered Machine) with an open and integrative approach. This competition takes place annually in Boston, USA, and it stimulates interdisciplinary groups to problem solving through genetically modified organisms. In line with this proposal, the team responsible for this project consists of undergraduate and graduate students from various institutes of the University of São Paulo. The blending of open approaches and such interdisciplinary groups contributes to the development of the research per se , ultimately impacting its quality and depth for the better. The project is also integrated with the SynBio Brasil community, actively engaged in promoting synthetic biology education, leading to a powerful impact on scientific awareness in Brazil.
Implementation
Plasmid Construction
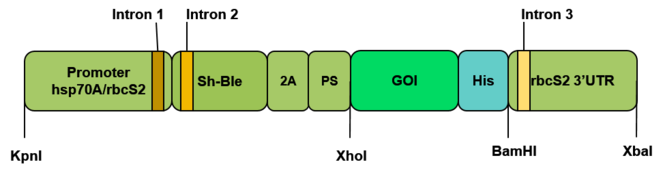
The vector pBluescript II (Thermo Fisher Scientific Inc.) will be used. The constructed cassette will be flanked by the restriction sites Kpn l at one end, and Xba l at the other. There are also two different restriction sites, Xho l and Bam HI, flanking the coding sequence of the desired protein. The codons of the proteins will be optimized for the expression in C. reinhardtii nucleus (
Cassette construction to be inserted in C. renhardtii nuclear genome for the expression of desired proteins. Promoter hsp70A/rbcs2: fusion of the promoters hsp70A and rbcs2 (
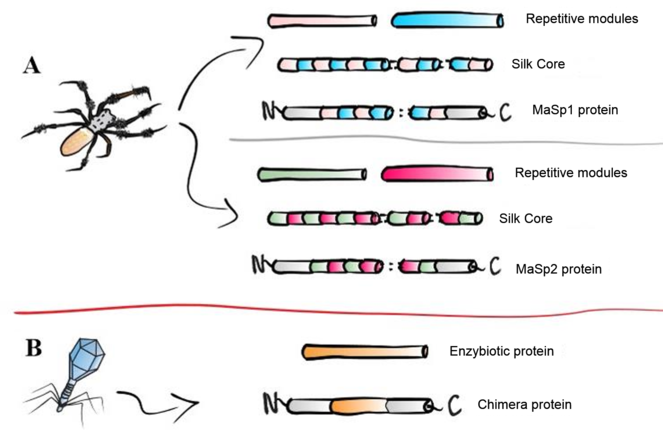
The native proteins MaSp1 and MaSp2 (Major Ampullate Silk Protein) were selected since they are the most studied proteins among the cob constituents. The antimicrobial enzymes were selected from a screening of the phiBIOTICS databank (
Desired proteins to be expressed in C. renhardtii.
NTD: N-terminal domain of MaSp 1. CTD: C-terminal domain of MaSp1. MRSA: Methicillin-resistant S. aureus. VRSA: Vancomycin-resistant S. aureus. VISA: Vancomycin-intermediate S. aureus. ORSA: Oxacillin-resistant S. aureus. CDS: Coding DNA sequence. MaSp1: Major Ampullate Silk Protein 1. MaSp2: Major Ampullate Silk Protein 2.
|
Protein |
Source |
Information |
Ref. |
|
MaSp1 |
Latrodectus hesperus (Black Widow) |
CDS fully sequenced |
|
|
MaSp2 |
Latrodectus hesperus (Black Widow) |
CDS fully sequenced |
|
|
NTD + MV-L + CTD |
Bacteriophage phiMR11 |
Effective against MRSA, VRSA e VISA |
|
|
NTD + LysK + CTD |
Bacteriophage K |
Effective against MRSA e VRSA |
|
|
NTD + Lysostaphin + CTD |
Staphylococcus simulans |
Effective against MRSA, ORSA e VISA |
|
C. reinhardtii cultivation
The C. renhardtii strain cc1690 will be obtained at the Chlamydomonas Resource Center. After going through the process of transformation, the ones identified to produce the protein of interest will be grown for protein production. The strains will be cultivated in TAP (Tris-Acetate-Phosphate) medium (
C. reinhardtii transformation
C. renhardtii cells will be grown in TAP medium until reaching the cell density of 3-6 x 106 cells/mL. The cells will be collected by centrifugation and resuspended in TAP enriched with 40 mM of sucrose reaching a cell density of 3-6 x 10 6 cells/mL. Then, 250 μL of the culture will be incubated with 300-1000 ng of plasmidial DNA, previously linearized through the digestion with Xba l and Kpn l for 5-10 min in cuvettes kept in an ice bath. An exponential electrical pulse of 2000 V/cm will be applied to the sample with an electroporation device, GenePulser XCellTM (BioRad, Hercules, CA). Capacitance will be adjusted to 25 mF and the resistance will not be regulated. After that, the cells will be incubated for 18 h in 10 mL of TAP/40 mM sucrose and plated in TAP/Zeocin solid medium (
Selection of mutants and identification of recombinant proteins
Recovered cells will be plated on TAP agar medium with increasing antibiotic concentrations (0.1, 2 and 5 μg/mL Zeocin). Candidate transformed colonies, displaying high Zeocin resistance, will be analyzed through PCR screening, and PCR positives colonies will be tested for protein of interest production by Western blot. Basically, the mutant cells are cultured as described above and fractions of the supernatant and cell lysate will be tested for the presence of the protein of interest. Cell lysis will be accomplished by sonication as described in the literature (
Western Blot
Samples of supernatant and total soluble proteins will be denatured by adding SDS-PAGE loading buffer (Laemmli) with β-mercaptoethanol, followed by incubation at 95 °C for 5 min. Proteins will be separated on 12% polyacrylamide gels at 120-150 V and transferred to nitrocellulose membrane at 200mA for 1h. Then they will be blocked in 5% solution of skimmed milk and the protein of interest will be probed with monoclonal mouse anti-His antibody. The membrane will then be washed 3 times with TBS-T (Tris-buffered saline with Tween 20 detergent) for 10 min and incubated with anti-mouse antibody conjugated with alkaline phosphatase, 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and nitroblue tetrazolium (NBT) substrates which forms an insoluble dark blue diformazan precipitate, allowing protein identification.
Recombinant proteins purification
Purification of the proteins of interest will be carried out by Nickel resin (Ni Superflow His60, Clontech®) following the manufacturer's instructions. The method is based on the affinity interaction with the hexa histidine tail, present in recombinant protein with the already mentioned resin. Basically, the sample is added to the column with the resin precharged with nickel ions, in which the proteins of interest containing histidine residues on its surface will be attached. Proteins not bound to the column will be washed out with the wash buffer, while the protein of interest is eluted with buffer containing 500 mM Imidazole.
Quantification of recombinant proteins
Quantification of purified proteins will be obtained via the Enzyme Linked Immunosorbent Assay (ELISA). Thus 200 μL of sample are incubated in 96-well plates at 37 °C for 30 min, then the solutions are removed, blocked in 5% solution of skimmed milk and the wells are washed 3 times with TBS-T (Tris-buffered saline with Tween 20 detergent) . Then, 200 μL/well of TBS-T solution of monoclonal mouse anti-His is added and incubated at room temperature for 2 h and washed as described above. A new TBS-T solution with anti-mouse monoclonal antibody conjugated with alkaline phosphatase is added, incubated at room temperature for 2 h and washed 3 times with TBS-T for 10 min. For the development, a freshly prepared solution of p-nitrophenyl phosphate is added and incubated in the wells for 30 min, and the plate is subsequently read in a plate reader at 405 nm.
Data analysis
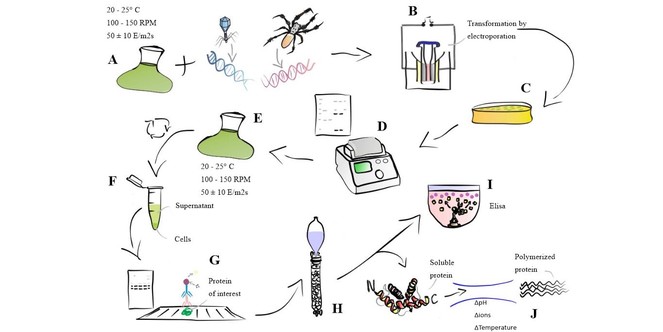
Results will be evaluated by analysis of variation (ANOVA) performed in Statistica software 10. Statistical significance will be evaluated by estimating the descriptive level (p) and the results will be considered statistically significant at p < 0.05 (confidence level of 95%) .The methods described above are shown in the flowchart in
Chronogram of execution
|
Month |
1 st |
2 nd |
3 rd |
4 th |
5 th |
|
Plasmid construction |
X |
||||
|
Chlamydomonas reinhardtii transformation |
X |
X |
|||
|
Transformed strains evaluation |
X |
X |
X |
X |
|
|
Cob polymerization evaluation |
X |
X |
|||
|
Results analysis |
X |
X |
X |
X |
X |
|
Wiki development |
X |
Experimental Flowchart. (A) Wild Cells incubated with built vectors. (B) Wild-cell transformation by electroporation. (C) Selection of mutants resistant to Zeocin. (D) Screening of antibiotic resistant cells by PCR. (E) Cultivation of PCR positive cells. (F) Fractions to be tested for the presence of recombinant proteins. (G) Detection of recombinant proteins present in the fractions by Western Blot. (H) Protein Purification. (I) Quantification via ELISA. (J) Spider silk polymerization reaction.
Acknowledgements
We would like to thank Nicolau de Almeida for reviewing the manuscript for the use of English.
Hosting institution
University of São Paulo.
Author contributions
All the authors contributed to this manuscript by reviewing and editing the final text, as well as either writing, doing background research or making figures.
Conflicts of interest
The authors report no conflict of interest.
References
-
Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains.PLoS Biology12(8):e1001921. DOI: 10.1371/journal.pbio.1001921
-
Blueprint for a High-Performance Biomaterial: Full-Length Spider Dragline Silk Genes.PLoS ONE2(6):e514. DOI: 10.1371/journal.pone.0000514
-
Spider Silk Capsules as Protective Reaction Containers for Enzymes.Advanced Functional Materials24(6):763‑768. DOI: 10.1002/adfm.201302100
-
Nanodevices for the immobilization of therapeutic enzymes.Critical Reviews in Biotechnology8551:1‑18. DOI: 10.3109/07388551.2014.990414
-
Uncorking the biomanufacturing bottleneck.Nature Biotechnology20(8):777‑779. DOI: 10.1038/nbt0802-777
-
Strategies to facilitate transgene expression in Chlamydomonas reinhardtii.Planta229(4):873‑883. DOI: 10.1007/s00425-008-0879-x
-
A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii+.The Plant Journal19(3):353‑361. DOI: 10.1046/j.1365-313x.1999.00526.x
-
Untangling spider silk evolution with spidroin terminal domains.BMC Evolutionary Biology10(1):243. DOI: 10.1186/1471-2148-10-243
-
Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii.Proc. Natl Acad. Sci. USA54:1665‑1669. DOI: 10.1073/pnas.54.6.1665
-
Polymeric materials based on silk proteins.Polymer49(20):4309‑4327. DOI: 10.1016/j.polymer.2008.08.006
-
Construction of modular tandem expression vectors for the green alga Chlamydomonas reinhardtii using the Cre/lox-system.BioTechniques43(3):324‑332. DOI: 10.2144/000112556
-
phiBIOTICS: catalogue of therapeutic enzybiotics, relevant research studies and practical applications.BMC Microbiology13(1):53. DOI: 10.1186/1471-2180-13-53
-
Recent Advances and Applications of Immobilized Enzyme Technologies: A Review.Research Journal of Biological Sciences5(8):565‑575. DOI: 10.3923/rjbsci.2010.565.575
-
Spider silks and their applications.Trends in Biotechnology26(5):244‑251. DOI: 10.1016/j.tibtech.2008.02.006
-
Efficient recombinant protein production and secretion from nuclear transgenes in Chlamydomonas reinhardtii.Journal of Biotechnology167(2):101‑110. DOI: 10.1016/j.jbiotec.2012.10.010
-
Transgenic microalgae as green cell-factories.Trends in biotechnology22(1):45‑52. DOI: 10.1016/j.tibtech.2003.11.003
-
Spider Silk: Ancient Ideas for New Biomaterials.Chemical Reviews106(9):3762‑3774. DOI: 10.1021/cr010194g
-
Evaluation of immobilized enzymes for industrial applications.Chemical Society Reviews42(15):6236. DOI: 10.1039/c3cs35511j
-
An Albumin-Derived Peptide Scaffold Capable of Binding and Catalysis.PLoS ONE8(2):e56469. DOI: 10.1371/journal.pone.0056469
-
Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron.The Plant Journal14(4):441‑447. DOI: 10.1046/j.1365-313x.1998.00145.x
-
Expression and assembly of a fully active antibody in algae.Proceedings of the National Academy of Sciences100(2):438‑442. DOI: 10.1073/pnas.0237108100
-
Chlamydomonas reinhardtii chloroplasts as protein factories.Current Opinion in Biotechnology18(2):126‑133. DOI: 10.1016/j.copbio.2007.02.001
-
Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials.PLoS ONE4(3):e4944. DOI: 10.1371/journal.pone.0004944
-
Immobilization of β-galactosidase from Escherichia coli onto modified natural silk fibers.Journal of Applied Polymer Science130(4):2923‑2931. DOI: 10.1002/app.39475
-
The Recombinant Phage Lysin LysK Has a Broad Spectrum of Lytic Activity against Clinically Relevant Staphylococci, Including Methicillin-Resistant Staphylococcus aureus.Journal of Bacteriology187(20):7161‑7164. DOI: 10.1128/jb.187.20.7161-7164.2005
-
Endotoxin removal from protein solutions.Journal of Biotechnology76:97‑119. DOI: 10.1016/s0168-1656(99)00185-6
-
L-asparaginase treatment in acute lymphoblastic leukemia.Cancer117(2):238‑249. DOI: 10.1002/cncr.25489
-
The microalga Chlamydomonas reinhardtii as a platform for the production of human protein therapeutics.Bioengineered Bugs2(1):50‑54. DOI: 10.4161/bbug.2.1.13423
-
Robust Expression and Secretion of Xylanase1 in Chlamydomonas reinhardtii by Fusion to a Selection Gene and Processing with the FMDV 2A Peptide.PLoS ONE7(8):e43349. DOI: 10.1371/journal.pone.0043349
-
Efficient Elimination of Multidrug‐Resistant Staphylococcus aureus by Cloned Lysin Derived from Bacteriophage φMR11.The Journal of Infectious Diseases196(8):1237‑1247. DOI: 10.1086/521305
-
A green light for engineered algae: redirecting metabolism to fuel a biotechnology revolution.Current Opinion in Biotechnology19(5):430‑436. DOI: 10.1016/j.copbio.2008.07.008
-
Immobilization strategies to develop enzymatic biosensors.Biotechnology Advances30(3):489‑511. DOI: 10.1016/j.biotechadv.2011.09.003
-
The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas.The Plant Journal21(2):121‑131. DOI: 10.1046/j.1365-313x.2000.00652.x
-
Enzyme immobilisation in biocatalysis: why, what and how.Chemical Society reviews42(15):6223‑35. DOI: 10.1039/c3cs60075k
-
Structure–function–property–design interplay in biopolymers: Spider silk.Acta Biomaterialia10(4):1612‑1626. DOI: 10.1016/j.actbio.2013.08.020
-
Oxidation of Glucose to Gluconic Acid by Glucose Oxidase in a Membrane Bioreactor.Applied Biochemistry and Biotechnology121:0149‑0162. DOI: 10.1385/abab:121:1-3:0149
-
Biopharmaceutical benchmarks 2014.Nature Biotechnology32(10):992‑1000. DOI: 10.1038/nbt.3040
-
Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae.Current Opinion in Biotechnology24(3):405‑413. DOI: 10.1016/j.copbio.2013.04.004
-
In vitro activity of recombinant lysostaphin against Staphylococcus aureus isolates from hospitals in Beijing, China.Journal of Medical Microbiology56(1):71‑76. DOI: 10.1099/jmm.0.46788-0
-
Hyper-production of large proteins of spider dragline silk MaSp2 by Escherichia coli via synthetic biology approach.Process Biochemistry51(4):484‑490. DOI: 10.1016/j.procbio.2016.01.006