|
Research Ideas and Outcomes : NIH Grant Proposal
|
|
Corresponding author: Arno Klein (arno@binarybottle.com)
Received: 11 Apr 2016 | Published: 19 Apr 2016
© 2016 Arno Klein.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Klein A (2016) A game for crowdsourcing the segmentation of BigBrain data. Research Ideas and Outcomes 2: e8816. doi: 10.3897/rio.2.e8816
|

|
Executive summary
The BigBrain, a high-resolution 3-D model of a human brain at nearly cellular resolution, is the best brain imaging data set in the world to establish a canonical space at both microscopic and macroscopic resolutions. However, for the cell-stained microstructural data to be truly useful, it needs to be segmented into cytoarchitectonic regions, a challenge no single lab could undertake. The principal aim of this proposal is to crowdsource the segmentation of cytoarchitectonic regions by means of a computer game, to transform an arduous, isolated task performed by experts into an engaging, collective activity of non-experts.
Keywords
BigBrain, brain imaging, histology, microstructure, neurons, crowdsourcing, game, gamify
Research & Related Other Project Information
Project Decsription
The principal aim of this proposal is to crowdsource the segmentation of brain histological data, specifically the cytoarchitectonic regions of the hippocampi of the human brain, by means of a computer game. Currently, only human experts perform reliable cytoarchitectonic labeling at a very slow and small scale, whereas we propose to enlist many human non-experts to engage in a distributed version of this task at a quick and large scale. By turning this arduous, isolated task into an engaging, collective activity, we hope to radically change the way anatomists approach segmentation/labeling.
To support this aim, we must (1) prepare expert (gold standard) labels to a subset of the hippocampal sections to evaluate crowdsourced results, and (2) aggregate the crowdsourced results to label the hippocampi. For our first exploratory aim, will explore how our approach generalizes to all other brain regions, and for our second exploratory aim, we will train a supervised learning algorithm on the crowdsourced results and evaluate how closely the automated approach matches human assessments.
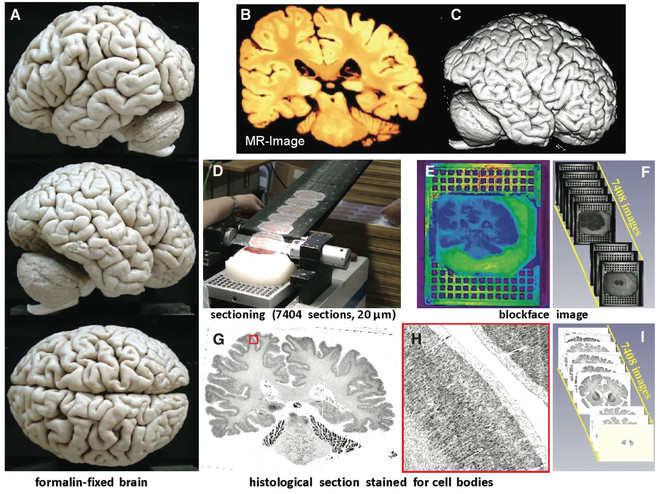
We will use the BigBrain, a high-resolution 3-D model of a human brain at nearly cellular resolution (20μm isotropic) based on reconstruction of 7,404 histological sections stained for cell bodies. The BigBrain is extremely important to the neuroscience community because it represents whole-brain histological data with accompanying magnetic resonance imaging data. It is the best brain imaging data set in the world to establish a canonical space at both microscopic and macroscopic resolutions. However, for the cell-stained microstructural data to be truly useful, it needs to be segmented into cytoarchitectonic regions, which makes it the perfect focus for this project. Labeling of the BigBrain will establish an ex vivo atlas, a common space for neuroimaging data whose labels will provide a consistent, convenient, and meaningful way to communicate, classify, and analyze biomedical research data set in that space.
Facilities & Other Resources
Sage Bionetworks
Laboratory: N/A
Clinical: N/A
Animal: N/A
High Performance Computing Resources: Sage Bionetworks uses a combination of scalable cloud-based storage and analytical computational resources and its own computational facilities. The cloud-based services are procured from Amazon Web services on a fee for service basis and provide a cost-effective solution to variable needs, technology upgrades and support. Sage Bionetworks develops and operates two software as a service platforms, Bridge and Synapse, as resources for the broader scientific community. Both these systems opperate on cloud-based infrastructure. Internal research projects also have access to the Sage Bionetworks high performance computing cluster, maintained through a partnership agreement with the University of Miami.
Additional servers used by Sage scientific staff are co-located at the Fred Hutchinson Cancer Research Center computing facilities. All networked file systems, databases, and home directories are backed up using Veritas software to a robotic tape library. Tapes are taken off-site each month for disaster recovery.
Additional facilities/resources :
Sage Bionetworks Bridge platform is designed to support the design and execution of adaptive clinical trials delivered though smartphone platforms. Participants in a study interact with the Bridge server through an app which is custom-designed for each particular study. The Bridge server provides a way to configure a study with a set of survey questions to ask study participants and way to schedule times for the various survey and app data collection tasks to run. Storing this configuration on the server allows researchers to dynamically adjust the study as data is collected without distributing new builds of the app to participants.
The Bridge server provides services for study participants to create an account, authenticate, and manage informed consent to participate in a research study. The app will periodically save survey and sensor measurements to the server; client-side SDKs help manage transmission of his data over intermittent mobile network connections. Study data is stored separately from readily identifying user account data so that study participants can be deidentified and data shared protecting their anonymity. Once deidentified, study data will be stored on Sage Bionetworks Synapse platform.
Sage Bionetworks Synapse platform will be used to support the complex, interrelated analyses described in this project. Synapse is an informatics platform for collaborative, data-driven science that combines the power of community-based modeling and analysis with broad access to large datasets, which will enable the development of more predictive computational models of disease. Synapse is built as a web service-based architecture in which a common set of services is accessed via different sets of client applications, including a web portal and integrations with multiple analysis environments.
Synapse is designed to allow users to bundle together and publish the relationships, annotations, and descriptions of files that may live in multiple locations. This may be files stored in Synapse own native storage location (Amazon S3), data living on local file systems, or code from GitHub. As long as the storage location is accessible via http/ftp it can be accessed through Synapse. Synapse includes analysis clients for users working in the R and Python programming languages, and a tool that runs at the program line of the Linux shell, providing basic functionality to interact with the system no matter what analysis tools the analyst would like to use. These tools allow users to query and load data, post results, and create provenance records directly from the command line. The output of any analysis can be stored in the Synapse Amazon S3, or externally and indexed in Synapse just as the underlying data. The Synapse web portal is an environment for sharing data, results, methods and tools, and is a place that enables the tracking of analysis steps and publication of analysis results to collaborators and eventually the broader community. The Synapse web portal will allow researchers to publish analysis results and track project information. A Provenance visualization tool will allow users to formally track the relationship among resources in the system and better document and communicate each analysis step.
The Synapse system is operated as a hosted service offering from Sage Bionetworks, requiring no installation of software or IT burden on the collaborating institutions. Researchers will be able to create accounts in the system and immediately use the system to collaborate.
At this time Synapse is hosted at http://synapse.org, and is being used to host a variety of bio-molecular data sets and analytical pipelines to curate this data. Sage Bionetworks has successfully used Synapse to support a number of large scale collaborative projects including open challenges in the predictive modeling of breast cancer survival and the TCGA Pan-Cancer working group.
Scientific Environment: Sage Bionetworks leases approximately 3,900 sq ft of office space on the Fred Hutchinson Cancer Research Center campus (FHCRC or the Center.) As part of the agreement with FHCRC, all Sage Bionetworks staff have full access to the Center’s research. Sage Bionetworks has a services agreement with FHCRC for facilities related logistics.
Centre de Recherches Interdisciplinaires (CRI)
Clinical: N/A
Animal: N/A
High Performance Computing Resources: N/A
Laboratory: Inserm U1001 lab in the Faculty of Medicine of the Paris Descartes University, working on Robustness and evolvability of life.
The lab offers the following facilities:
Microscopy:
- Axiovert 200 M and Axioplan 2 and Observer Z1 from Zeiss
- Eclipse Ti, Eclipse E200 from Nikon
- MetaMorph (software)
Microfluidics:
- SEM
- Chemostat
llumatool tunable lighting system
Microplates reader:
- iEMS (Labsystems)
- Victor3 (Perkin Elmer)
- Infinite (tecan)
Research in this lab, headed by F. Taddei and A. Lindner, is focused on studies into key issues such as aging on one hand and the origin of cooperation on the other, using state-of-the-art systems and synthetic biology approaches, including computer modelling, wet-lab experiments, robotics and nano-fabrication methods.
Additional facilities/resources :
The MOOC Factory
A platform to create MOOCs to support the CRI’s own education programmes but also to disseminate its interdisciplinary approaches to a larger public. In the factory, online courses focused on practice and experimentation are developed by innovators who wish to create courses for the general public, with a special emphasis for teachers, parents and children.
OpenLab (DIY research FabLab)
A place where students and entrepreneurs can prototype, code, hack and take part to the creation of connected objects and captors, around a multidisciplinary team.
The Lab has all the basic tools to prototype rapidly captors and interface them with web apps. Students can also use 3D printers, a laser cutter and a digital milling machine. A DIY-bio lab is also hosted at Cochin.
The OpenLab is a key resource to build and nurture an extended and diverse CRI community of bright minds. In less than two years, the OpenLab has attracted more than a hundred candidates and has selected 15 promising teams of young entrepreneurs on topics as diverse as quantified self, green mobility or open health.
See the list of selected start-ups (more information: http://lopenlab.org/projects-startup/promo-2-2015/ and http://lopenlab.org/projects-startup/promo-1-2014/).
Education experience
The CRI offers an Interdisciplinary curriculum, which spans the range from undergraduate (one Bachelor programme: Frontières du vivant) through masters (2 programmes: Interdisciplinary Approaches to Life Sciences and EdTech) to PhD studies in the fields of interdisciplinary life sciences and learning sciences at Sorbonne Paris Cité, one of France's major university consortiums bringing together four Paris universities and four higher education and research institutes. The CRI hosts programmes for about 70 undergraduate students and 170 graduate students per year, in around fifty associated laboratories in the best French and international institutions, including the Rockefeller Institute, EMBL, Peking University, Harvard University, and Imperial College London. The academic programmes are supervised and evaluated by an international scientific committee.
Scientific Environment:
The CRI leases approximately 3,000 square meters in the heart of Paris, in Le Marais. These dedicated facilities host visiting professors, a variety of courses, and many student clubs. They are complemented by 350 square meters of labs in the Institut Cochin.
The CRI organises special programmes like the Leadership programme, which brings top-notched young researchers in Life Sciences to develop and implement innovative educational projects. During the three days of this programme, everybody present at the CRI can take part and interfere with the selected researchers.
The CRI also host Scientific Clubs created by students on a wide range of subjects of their choice such as interdisciplinarity, innovation, social and environmental development, science and society… The clubs give students the opportunity to get involve in an extra-curricular activity that matches their interests and adds creativity, collaborativity and design thinking to their skill set. Current active clubs include WAX (aiming at creating and developing innovative and collaborative tools to make science fun, accessible to all and useful), Fabelier (a lab to create new things with/for the web) and Gamelier (for those who want to meet up to make games together, with a focus on educational and scientific games).
iGEM (international genetically engineered machine): the first French synthetic biology team for the MIT-sponsored iGEM competition. In this club, the CRI students have collected many awards through the years. In 2013, they were honoured with the Grand Prize at the World Championship Jamboree.
All the CRI activities, whether they are primarily focused on Education, Research or Entrepreneurship, attract and build up an exceptional community of bright minds inhabited by a common vision that knowledge sharing and open collaboration are important keys to our future. The diversity, quality and benevolence of the ever growing CRI community makes this place a unique resource to forge serendipity and unlock the unexpected.
The CRI hosts the Liliane Bettencourt Interdisciplinary Programme, which spans the range from undergraduate through masters to PhD studies. This 200-strong group comprises students from all the sciences, who carry out research in the best French and international institutions, including the Rockefeller Institute, EMBL, Peking University, Harvard University, and Imperial College London. The academic programmes are supervised and evaluated by an international scientific committee. CRI has initiated projects such as the MIT award-winning Paris synthetic biology iGEM team and interdisciplinary approaches to web sciences, Fablab hacker space. Researchers at the CRI are involved in projects at the interface of science, education and society, ranging from the Paris Montagne Science Festival for young children to outreach programmes for high school students from disadvantaged environment (Science Ac’87) and graduate students. CRI organizes Learning through Research workshops for students and lecturers across the world (Europe, China, Indonesia) and has launched the WISER-U as an international platform for students and scientists to design and develop new technologies for sharing ideas and creating projects together. Research at the CRI “Evolutionary Systems Biology” INSERM team, headed by F, Taddei and A. Lindner is focused on studies into key issues such as Aging on one hand and the origin of Cooperation on the other, using state-of-the-art systems and synthetic biology approaches, including computer modelling, wet-lab experiments, robotics and nano-fabrication methods.
The CRI’s dedicated facilities host visiting professors, a variety of courses, and many student clubs. It is host to Laboratory facilities of the U1001 unit of the French National Institute of Health & Medical Research (INSERM) in Paris-Descartes University’s Medical School; The OpenLab, a prototyping workplace and projects accelerator created to lower the entry barriers of technological and social innovation for students, scientists, designers, engineers, and entrepreneurs alike; The Game Lab, a place for students and researchers to learn about game creation and to develop their scientific games; The MOOC (Massive Open Online Courses) Factory, where online courses focused on practice and experimentation are developed by innovators who wish to create courses for the general public, with a special emphasis for teachers, parents and children.
McConnell Brain Imaging Center (BCI) at McGill University
About the Center
The McConnell Brain Imaging Centre ("The BIC") is a multidisciplinary research centre dedicated to advancing our understanding of brain functions and dysfunctions and the treatment of neurological diseases with imaging methods. It is also one of the largest, multimodal brain imaging service platforms worldwide (largest in Canada), serving a community of 100+ investigators and generating a volume of more than 3,500 research scans per year in 7 imaging cores: MRI, PET, SPECT, MEG, EEG, etc.
Our Centre is also renowned for developing and distributing software applications and for having created multiple spin-off biomedical imaging companies.
Core Facilities
The unique platform infrastructure of the McConnell BIC is available and accessible to all researchers.
We offer one of the largest brain-imaging research and service platform worldwide. All imaging cores are hosted under the unique roof of the Montreal Neurological Institute (MNI) and hospital. Every year, the McConnell BIC scans >3,500 research participants with a multimodality of 7 research-dedicated scanners for humans and animal models.
University of Pennsylvania
Penn is home to a diverse body of over 20,000 students and over 4,000 faculty in its 12 leading graduate and professional schools. Penn’s schools are located on a compact campus, the geographical unity of which supports and fosters its multidisciplinary approach to education, scholarship, and research. Research and research training are substantial and esteemed enterprises, bolstered by an annual University budget of $6 billion. Penn’s 165 research centers and institutes bring together researchers from multiple departments, schools, and disciplines, and interdisciplinary collaboration is a key theme for Penn’s academic enterprises.
Penn and the Perelman School of Medicine provide a truly outstanding intellectual environment in which to conduct the research we propose. The School of Medicine, and the Department of Radiology in particular, are consistently among the top five recipients of biomedical research funding in the nation. Virtually any kind of biomedical research is represented at Penn, and few other institutions offer a comparable breadth and depth of expertise and resources. This includes a wide range of imaging resources and expertise, including MRI, CT, 3D tomosynthesis, PET, 3D ultrasound, histology, and confocal microscopy.
An ongoing initiative to consolidate research in neuroimaging and brain function on the Penn campus is entering its second phase, and will result in nearly all neuroimaging investigators being located in newly renovated space centrally located on campus. By the end of 2015, Dr. Yushkevich’s laboratory will be located in the Richards and Goddard Laboratories Buildings, a designated National Historic Landmark designed by the renowned architect Louis Kahn to facilitate research interactions. Although the Penn campus is already very compact, consolidating the many faculty members involved in neuroimaging research will facilitate broader participation in projects, seminars, workshops, and interdisciplinary training, and will provide an outstanding environment for trainees to interact with faculty and with each other.
The primary site for this project will be the Penn Image Computing and Science Laboratory (PICSL) in the Department of Radiology. PICSL has an established track record of developing novel methodology for biomedical image analysis, disseminating methodology through open-source software, and applying it to relevant clinical problems through strong collaborations with clinical researchers at Penn and beyond. PICSL is home to over 20 graduate students, postdoctoral fellows and research staff. In summer 2015, PICSL will move to occupy a whole floor in the D tower of the Richards building, and will be steps away from the laboratories of leading neurologists and neuroscientists.
Computing Resources
Computer Hardware and Software Resources
The computing resources for this project will be available through the Neuroscience Neuroimaging Center (NNC), a NINDS P30 Institutional Center Core. John Detre is the PI of the NNC, and Paul Yushkevich is the PI of the Neuroinformatics Core, which maintains the NNC computing resources. The NNC maintains a state-of-the-art data processing facility with distributed computing, data storage and archiving, integration of neuroimaging data analysis environments, and provides system administration support. The NNC high-performance computing cluster consists of
576 dedicated compute cores, from 21 compute nodes with dual 8-core Intel Xeon E5-2450 2.10GHz CPUs and 64GB RAM, and 30 compute nodes with dual 4-core Intel Xeon 2.83GHz E5-440 CPUs with 16GB RAM
A dedicated 16-core head node (running Rocks and SGE) manages cluster operations
Three dedicated file servers managing high-speed (6GB/s and 4GB/s) RAID-6 devices for over 200TB of formatted storage.
A high-speed 10GbE internal cluster network and a Gigabit external network connection.
A dedicated tape backup system with 100TB capacity.
The cluster is located in a University-run commercial-grade server room with redundant power supplies; UPS power backup systems; fire suppression system; and 24-hour restricted access and security.
Additionally, each investigator will have a personal workstation (Apple iMac or similar) with gigabit connection to the University of Pennsylvania Health System (UPHS) intranet, as well as fast Internet access. A range of software is available on the investigators’ workstations and on the HPC cluster, including multi-platform software development tools, scientific computing packages, functional and structural MRI analysis frameworks; statistical packages, word processing and presentation applications, etc.
Resources for Open-Source Software Development and Dissemination
PICSL has recognized expertise and a strong track record of open-source scientific software development. PICSL was one of the founders of the National Library of Medicine Insight Toolkit (ITK) and is a major contributor to the most recent ITK version 4. PICSL leads the development of other significant open-source tools, including interactive image segmentation tool ITK-SNAP, deformable image registration package ANTS/SyN, and diffusion MRI analysis package DTI-TK. All three of these open-source applications tools are used widely by researchers, and are estimated to have thousands of users. This project will take full advantage of the software development infrastructure and expertise at PICSL. These best practices include hosting source code, documentation, and Wikis on public repositories such as GitHub and SourceForge. Concurrent development by multiple programmers is facilitated by the use of Git software. Cross-platform nightly builds are performed on a dedicated server that contains multiple VMware Fusion virtual machines running different versions of Windows, MacOS, and Linux operating systems. These virtual machines are automatically activated at night to compile the latest source code in the repository, perform automated testing, and upload binary executables to a public file download area. The results of the compilation and testing are sent to a web-based dashboard. This build process relies on open-source CMake, CTest, CPack and CDash tools from KitWare, Inc.
Office Resources
PICSL currently occupies 2,000 ft2 of space on Penn campus, and in the summer 2015 will move to a larger 2,400 ft2 space in the renovated Richards building. Each investigator will have office space in this building, along with shared office and open space for trainees and students. Michele Haines, the dedicated administrative assistant for PICSL, will provide administrative support for the project.
Resources for Medial Temporal Lobe Mapping Research
Penn Memory Center (PMC) and Penn Frontotemporal Dementia Centers (PFDC) Cohorts
Building the combined in vivo / ex vivo MTL atlas in Aim 1 involves retrospectively accessing and analyzing in vivo MRI data from the records of patients evaluated at the Penn Memory Center (Dr. Wolk is the Assistant Director) and participants in imaging research studies conducted by both the Penn Memory Center and the and Penn Frontotemporal Dementia (FTD) Center, which is directed by Dr. Grossman.
The Penn Memory Center (PMC) is a single, unified Penn Medicine source for those age 65 and older seeking evaluation, diagnosis, treatment, information, and research opportunities related to symptoms of progressive memory loss, and accompanying changes in thinking, communication and personality. The PMC is a National Institute on Aging-designated Alzheimer's Disease Center (ADC). It is one of only 30 such sites in the nation. ADC designation is earned by leading universities and medical institutions offering state-of-the-science diagnosis, treatment, research, and more for individuals with Alzheimer’s disease, mild cognitive impairment (MCI) and other age-related progressive memory disorders. Research is a chief mission of the PMC. The PMC is located in the 2 South wing of the UPHS’ new Perelman Center for Advanced Medicine (PCAM). The PMC is a multidisciplinary clinical and clinical research center providing state-of-the-art diagnosis and care for adults with cognitive disorders. It is designed for efficient research and clinical patient flow, comfort, and safety. The PMC currently includes six faculty clinician-researchers from 3 departments (psychiatry, neurology, medicine), two fellows, one resident, a nurse and other staff who provide patient care and conduct research studies. The PMC performs cutting edge patient-oriented clinical research. Currently, the PMC is conducting five NIH funded studies, 4 pharmaceutical industry clinical trials, and two private foundation studies of cognition in normal adults, mild cognitive impairment, Alzheimer’s disease and vascular cognitive impairment.
The Penn FTD Center is a state-of-the-art research center that brings together leading experts in neuropsychology, neuroimaging, clinical care, biomarker and cognitive neuroscience in an effort to improve the diagnosis, treatment, and care for individuals with FTD. In 2012, the Center evaluated over 500 patients. Currently, 240 patients participate in research projects that involve multimodal clinical, imaging, biofluid biomarkers, and extracted DNA data. The Penn FTD Center works in close collaboration with the Center for Neurodegenerative Disease (CNDR) Neuropathology and Genetics Cores and with other neurodegenerative disease centers at the University of Pennsylvania. These include the Penn Memory Center, the Amyotrophic Lateral Sclerosis (ALS) Center, and the Morris K. Udall Center of Excellence for Parkinson's Disease Research.
Both centers operate multiple research neuroimaging studies, and the oblique coronal T2-weighted scan described in the Research Strategy is routinely acquired since 2010. The sequence was also incorporated into the clinical MRI protocol at the Penn Memory Center in 2011. As of January 2015, the Penn FTD Center had collected such scans in over 500 subjects, and the Penn Memory Center had collected them in over 800 subjects. Approximately 34% of the subjects participating in the T2-weighted scans consent to donate their brain to autopsy (50% of subjects at the Penn FTD Center and 60% of subjects involved in the National Alzheimer’s Coordinating Center (NACC) study at the Penn Memory Center, who represent ~35% of all scans at the Penn Memory Center). Autopsies are performed by CNDR pathologists and coordinated by the CNDR Brain Bank.
Center for Neurodegenerative Disease Research (CNDR) Brain and Biospecimen Banks
Tissue specimens (n=30) for the in vivo / ex vivo atlas proposed in Aim 1 will be obtained through a collaboration with the Center for Neurodegenerative Disease Research (CNDR). The CNDR brings together researchers investigating the causes and mechanisms of neurodegenerative diseases that occur more frequently with advancing age, including AD, Parkinson's disease and other Lewy body diseases, frontotemporal degeneration diseases, amyotrophic lateral sclerosis and other motor neuron diseases. Its core and affiliated research programs encompass basic cell and molecular biology of protein misfolding and neurodegeneration, neurogenetics, neuropathology, biomarker discovery and validation, translational medicine and drug discovery, clinical research programs including the Penn Memory Center / Alzheimer's Disease Center, and a database to integrate data from all of these programs on a common platform.
The CNDR established a biosample collection in 1989 to procure brains, cerebrospinal fluid, DNA, and plasma and serum from normal aged controls and patient subjects with neurodegenerative disorders. All specimens are tracked and linked with demographic and clinical data in a database and all samples and data are coded and de-identified to maintain patient confidentiality in accordance with HIPPA standards. Organ/tissue processing is optimized for multiple uses including confidentiality in accordance with HIPPA standards. All brain autopsies and diagnostic neuropathological examinations are performed in-house, under the direction of Dr. Trojanowski. Fixed and frozen tissues are currently available for research studies from more than 1,400 individuals. In addition, cerebrospinal fluid is available from >850 subjects and DNA/plasma/serum from >1,400
Small Animal Imaging Facility (SAIF)
Ex vivo MRI studies in Aim 1 will be carried out at the SAIF, which provides multi-modality radiological imaging and image analysis for cells, tissues, and small animals. The SAIF combines state-of-the-art instrumentation and a nationally recognized staff to assist investigators with a wide range of imaging based experimental approaches. The SAIF currently provides a comprehensive suite of imaging modalities including: magnetic resonance imaging (MRI) and spectroscopy (MRS), optical imaging (including bioluminescence, fluorescence, and near-infrared imaging), computed tomography (CT), positron emission tomography (PET), single photon emission computed tomography (SPECT), and ultrasound (US). In addition, dedicated housing is available for mice and rats undergoing longitudinal imaging studies. Ancillary facilities and resources of the SAIF are devoted to chemistry, radiochemistry, image analysis and animal tumor models.
Ex vivo MRI will utilize a 9.4 Tesla (Varian, Inc., Palo Alto CA) 31 cm horizontal bore MR system (B100 John Morgan Building) equipped with a 21 cm ID 25 gauss/cm and a 12 cm ID 40 gauss/cm gradient tube and interfaced to an Agilent DirectDrive console. A custom M2M quadrature transmit/receive body coil with inner diameter of 35mm and a long z field (supporting B1 field of 80 mm FOV along the length of the specimen) was purchased in 2010 specifically to support human MTL tissue imaging. A number of other coils are also available.
Tissue preparation for imaging studies in Aim 1 will take place in a fully equipped animal surgery room adjacent to the Varian 9.4 Tesla MRI scanner in 101A John Morgan building. The scanner room also provides dedicated storage space our MTL tissue specimens.
CHOP Pathology Core
The scanning of histology slides in this project will be carried out at the Pathology Core Laboratory at the Joseph Stokes Jr. Research Institute in the Children’s Hospital of Philadelphia. In addition to a full range of histopathology services, the Core provides sophisticated imaging instrumentation for high-resolution whole slide scanning and virtual microscopy (ScanScope from Aperio). The PI has already collaborated with the CHOP core in the Phase 1 of the project. In the current proposal, the histology preparation and staining work will be carried out by through the subcontract to the Insausti group at UCLM, whose techniques are highly optimized for anatomical and pathological imaging of the MTL. However, the ScanScope at the CHOP Core provides the best image quality and resolution, and will continue to be used in the proposed project.
MRI Resources
As of the summer 2015, the MRI facilities at Penn will include two research-dedicated 3 Tesla Siemens Prisma scanners and a Siemens 7 Tesla whole-body research MRI system. These scanners are operated by the Center for Advanced Magnetic Resonance Imaging and Spectroscopy (CAMRIS), which is responsible for maintenance, safety and staffing of human research MRI systems in the Department of Radiology. Maintenance includes service, upgrades and cryogen refills. A local Siemens engineer is available to address malfunctions. A CAMRIS committee reviews applications for MRI research studies on the Siemens systems for safety and feasibility and provides approval. CAMRIS collects an hourly usage fee, currently $500/hr, to cover the cost of scanner use, including maintenance and staffing by a certified MRI technologist. Additional support for MRI studies will be available from the Data Acquisition Core of the P30-funded Neuroscience Neuroimaging Center. This Core provides technical support for magnetic resonance and optical imaging protocol development and implementation, imaging quality assurance, and for implementing approaches for ancillary methods.
Proposed MRI imaging for the aging study in Aim 2 will be carried out on a Siemens 7 Tesla whole-body research MRI system with a 32-channel head coil is located in the basement of the Stellar Chance Building on the School of Medicine campus, a minutes walk the Richards Building. This system also includes RF coils for 31P NMR spectroscopy and a custom hybrid head/neck coil for arterial spin labeling. A parallel transmission system is also available and can be used in conjunction with an 8-channel transmit/receive array. An outboard GPU processor has been interfaced with the 3T and 7T systems to allow rapid image reconstruction for multiband EPI and other high-throughput imaging.
Amyloid PET scanning with the F18 Florbetapir tracer in Aim 2 will take place at the Positron Emission Tomography (PET) Center at the University of Pennsylvania, one of the first PET Centers in the country. It was established in 1975 as a research facility for the study of human brain function. It continues to be a leading biomedical research institution with a large program in basic and clinical PET research. More than 5,000 PET studies are performed by the PET Center each year.
The PET Imaging Facility has four whole body PET/CT scanners, one Philips Gemini TF in the Hospital of the University of Pennsylvania and Philips Gemini TF Ingenuity, Philips Gemini TF BB, and Siemens mCT scanners in the adjacent outpatient building. Separate rooms are available for patient preparation and injection of radiopharmaceuticals. In addition, there are nearby hot labs for preparing and extracting doses and a blood lab for sampling and counting. The Philips Gemini TF Ingenuity will be used for this study. It has an open design that allows patients to see outside the scanner, which improves patient acceptance. It has an 18-cm axial FOV and 70-cm diameter gantry aperture. The intrinsic spatial resolution is 4.8 mm, with energy resolution of 12% for good scatter rejection, and very good system timing resolution of 500-550 ps, which enables time-of-flight (TOF) measurement for improved image quality and data quantitation. The TOF information along with physical data corrections (e.g. scatter, attenuation) are included in the system model of the iterative ML-EM list-mode algorithm which is used for image reconstruction. Attenuation correction and anatomical registration are provided by the CT scanner. The Philips Gemini TF Ingenuity is shared 50/50 between research and clinical examinations. Daily quality control is performed and system performance is maintained by the Physics Instrumentation Group in the Nuclear Medicine division.
A cyclotron and radiochemistry facility provides space for the JSW, BC 33015 cyclotron, and its accessories including targets for production of short-lived radionuclides as well as a radiochemistry laboratory for chemical syntheses by which product radionuclides are incorporated into trace molecules. The Facility includes: 1) Cyclotron vault, 2) Cyclotron maintenance workshop, 3) Control room, 4) Radiochemistry laboratory, 5) Mechanical room, 6) Power supply room, 7) Counting room.
Equipment
N/A
Specific research plan
Specific aims
We have entered an age of unprecedented scale, resolution, and accessibility of digitized biomedical image data. The BigBrain (
Our proposal joins the expertise of the following people: Dr. Arno Klein (Sage Bionetworks) has expertise in brain image processing and has led manual and automated human brain image labeling projects; Dr. Alan Evans (McGill University) helped to create the BigBrain and leads a team interested in making it more useful and more accessible, Dr. Paul Yushkevich (University of Pennsylvania) is a leader in hippocampal labeling of histology and MRI data; Dr. Jesse Himmelstein (Centre de Recherches Interdisciplinaires) creates video games for research and education and makes online platforms for sharing games; and Amy Robinson directs the EyeWire project.
Aim 1: To evaluate non-expert labels, experts will label a small subset of BigBrain hippocampal data. To provide training and to evaluate results of a crowdsourced task, it is necessary to establish gold standards. We will have experts manually label a portion of BigBrain hippocampal regions to establish these gold standards.
Aim 2: To scale up labeling efforts, we will break the labeling problem into smaller tasks within a computer game. Just as the EyeWire game breaks data into small cubes within which isolated tasks are performed by non-experts, we will break BigBrain data into smaller pieces to make labeling a parallel, distributed effort. To engage non-expert labelers, we will map individual tasks to elements of a game that a human would enjoy playing. If this game fails to engage sufficient numbers of players, we will enlist players through Amazon’s Mechanical Turk.
Aim 3: Aggregate the results of game play to label the BigBrain. We will combine the thousands of results of playing the game to establish labels for the BigBrain hippocampal regions.
Exploratory Aim 1: Assess the potential for extending the game to all brain regions. The hippocampus is an important region to label, but ultimately we want to label the entire brain, so we will explore how our approach generalizes to all other brain regions.
Exploratory Aim 2: Train a supervised learning algorithm on the crowdsourced results. We propose to radically scale up our ability to label imaging data by training a supervised learning algorithm on the crowdsourced labels, and evaluate how well we can automate labeling.
Research strategy
(A) SIGNIFICANCE
The importance of segmenting the BigBrain:
The BigBrain (
Delineation of regions on the BigBrain, whether obtained from a manual or automated approach, will help improve statistics in probabilistic maps of neocortical and subcortical areas, as well as fiber tracts (although the stain used was for cell bodies, some fibers can be followed in areas of high contrast).
These cytoarchitectonic maps can be warped to a common reference frame and used as a topographical reference for anatomical localisation of activations observed in functional imaging studies, or as a high-resolution 3-D neuroanatomical guide (atlas) for microsurgery.
Labeling the cytoarchitecture of the human brain requires great expertise acquired over many years of labeling. It is a slow and tedious process; it would take an expert many years to draw all of the major cytoarchitectonic boundaries for the 7,404 sections of the BigBrain, and unless one could convince many such experts to do it, the necessary inter-rater reliability statistics of consistency cannot be measured. In other words, it will be a long time before we see the cytoarchitectonic boundaries of the BigBrain comprehensively labeled by experts, diminishing the tremendous potential that this data set has to offer. In this project, we propose to overcome the daunting challenge of labeling BigBrain cytoarchitectonic regions through crowdsourcing.
Labeling the hippocampus:
The medial temporal lobe (MTL) is a complex brain region of enormous interest in research on memory, aging, psychiatric disorders, and neurodegenerative diseases. Within the MTL, the subfields of the hippocampus (cornu Ammonis fields CA1-CA4, dentate gyrus, subiculum) and the adjacent cortical subregions of the parahippocampal gyrus (entorhinal cortex, perirhinal cortex, and parahippocampal cortex) are understood to subserve different functions in the memory system (
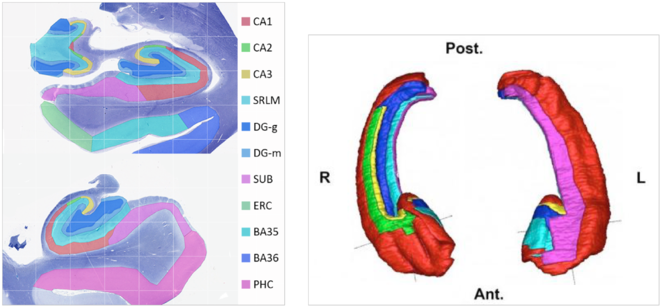
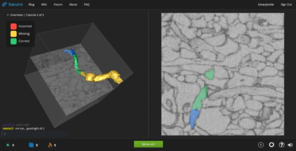
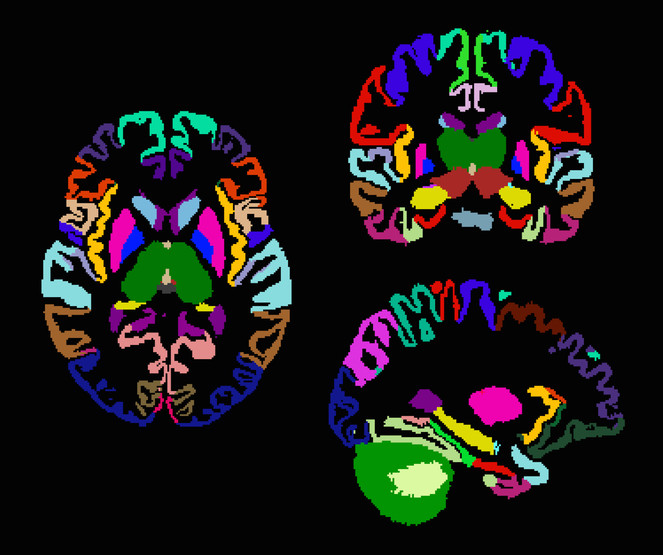
Dr. Yushkevich (University of Pennsylvania) is a world expert in hippocampal segmentation and works with hippocampal image data at both scales reflecting BigBrain modalities - microstructure and macrostructure (see
Example hippocampal subfield labels.
Left: Example of hippocampal subfield labeling. Right: The first work demonstrating hippocampal subfield labels in MRI space that are derived from ground-truth histological imaging (
Computer games for solving scientific problems:
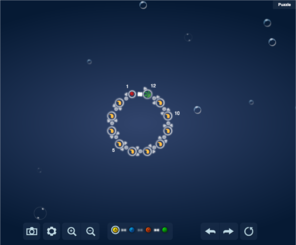
There has been increasing interest in and attention given to “citizen science” initiatives to crowdsource the solving of scientific problems through what is sometimes termed “human computation.” Compared to computers, humans currently excel at face and pattern recognition, object identification, abstract symbol processing, etc. Examples of recent scientific computer games which have attracted many players include Foldit and EyeWire (
Screenshots of popular scientific games.
FoldIt, Nanocrafter, and EteRNA tutorials are important for training and evaluating players and for providing scientific context, so that the player is further motivated to play. EyeWire has nearly 200,000 players who build neural connections from mouse scanning electron micrographs.
b: Nanocrafter tutorial
c: EteRNA tutorial
d: EyeWire tutorial
e: Eyewire neural reconstruction with scoring leaderboard
Foldit is an online puzzle video game where the objective is to fold the structure of selected proteins using various tools provided within the game. Foldit’s players (now estimated at 240,000 people (
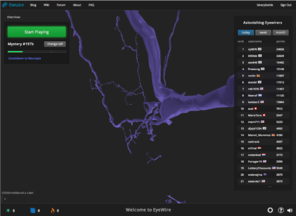
EyeWire is a game to help map neural connections in the visual cortex of a mouse brain played by nearly 200,000 people worldwide. The player does this by linking together computer-rendered, 3-dimensional segments of tissue extracted from scanning electron microscope sections to make neural processes that start from one face of a small cube of data to another face of the cube. By breaking the problem down from something currently computationally intractable (segmentation of interconnected neurons from SEM data) to one that is not only tractable but engaging for players (building a scaffold to escape a cube), they are able to crowdsource a very difficult problem. Likewise, the primary objective of our project is to crowdsource the segmentation of brain image data, in our case the cell-stained histological sections from a human rather than SEM sections from a mouse.
HTML5 technologies for browser-based scientific games:
The Web browser is doubtlessly the most widespread and cost-effective way to reach large numbers of potential players of scientific games. And yet, only a few years ago, technical difficulties prevented developers from delivering interactive and computationally intensive experiences over the Web. With the increasing uptake of the HTML5 standard by popular Web browsers, Web applications can now include 3-D graphics (WebGL), high-fidelity sound, and real-time communication with servers, all without browser plugins that require installation. Even better, many of these standards are being adopted by mobile Web browsers, meaning that the same application is available directly on a player’s smartphone or tablet.
The Centre de Recherches Interdisciplinaires (CRI) developed two technologies specifically created for scientists to take advantage of HTML5 technologies for games. RedWire (
Screenshots of a RedWire game and its RedMetrics game analytics.
Hungry Animal Train is a game created at the CRI Game Lab to study time estimation among children with autism. It was created entirely within RedWire, and publishes data about its use to the RedMetrics game analytics service, where researchers, developers, parents, and other interested partners can filter and download the data for statistical analysis.
b: Analytics screenshot
RedMetrics (
(B) INNOVATION
Aim 1, while focusing on traditional neuroanatomical methods for segmenting brain cytoarchitecture, is nonetheless innovative since it will modify a hippocampal labeling protocol for use with ex vivo hippocampal data, and will make use of these data as gold-standard labels for evaluation and scoring of players of a computer game.
Aim 2 is innovative since it will be the first instance of crowdsourcing cytoarchitectonic processing or analysis, and that too by individuals who have no neuroanatomical expertise (in contrast with Aim 1). Moreover, this crowdsourcing will take the form of a computer game, and will be the first to deal with cytoarchitectonic data. Players’ repeated segmentations will also be used to generate intra- and inter-rater reliability statistics for cytoarchitectonic segmentation, which will complement existing data on inter-brain anatomical variation of cytoarchitectonic boundaries (
Aim 3 will aggregate results from the crowdsourcing above, and will constitute the first time cytoarchitectonic boundary delineation will have been undertaken on small portions of the image for which (macrostructural) anatomical context has been removed. Another innovative contribution will be the software written to aggregate and reconcile tens of thousands of drawn curves to construct 3-dimensional boundary surfaces.
Exploratory Aim 1 is innovative in that it proposes to generalize a game-based approach tailored to a specific domain (the hippocampus) to a larger context (the entire brain).
Exploratory Aim 2 proposes the use of crowdsourced segmentations to train a machine learning algorithm to perform the same task. If this algorithm performs well, this will introduce a new and powerful approach to inferring cytoarchitectonic boundaries of brain regions.
(C) APPROACH
Aim 1: To evaluate non-expert labels, experts will label a small subset of BigBrain hippocampal data.
Dr. Yushkevich will oversee the manual segmentation of BigBrain hippocampal sections for use as a gold standard to evaluate each player’s accuracy and consistency, and will use this information to weight player contributions accordingly. Manual segmentation of the entire hippocampus at the resolution of the BigBrain is beyond the scope of this proposal. The UPenn team will label the left and right hippocampal regions in a subset of 60 slices each, taken in the coronal plane with approximately 1mm intervals. This represents 2% of the total number of slices spanning the hippocampus. We will store and release these segmentations and their boundaries as a publicly accessible project in Sage Bionetworks’ Synapse platform.
Segmentation Protocol :
The segmentation protocols by Ding and Van Hoesen (
Boundaries:
Boundaries can be defined either by a difference in the size/shape/density of cells, or in the presence/absence of layers. The hippocampus is considered more primitive cortex and doesn't have the distinct laminar appearance of the cortex. However, parts of the head of the hippocampus do appear this way, and this is one of the ways an expert uses to differentiate regions. The head of the hippocampus also has multiple classes of cells within each subfield. For example, CA3 cells appear quite different in the uncal, vertical, and typical regions (
Substructures :
The segmentation protocol will label the following hippocampal subfields: Cornu Ammonis (CA) fields CA1, CA2, and CA3; Dentate gyrus hilar region (DG:H), which is also referred to as CA4 by some authors, and the dentate gyrus proper (DG); subiculum (SUB). The hippocampal layers stratum radiatum and stratum lacunoso-moleculare will be assigned a separate label (SRLM), which correspond to the prominent hypointense band in MRI images of the hippocampus.
Operationalization :
Segmentation will be performed by John Pluta, who has over 5 years of expertise in hippocampal segmentation including ex vivo MRI and histology. He will use the HistoloZee tool (see
Reliability :
Inter-rater and intra-rater reliability for the protocol will be established. A set of 10 slices will be used for this purpose. John Pluta will resegment these slices after 1 month or more delay to establish intra-rater reliability. Laura Wisse will perform separate segmentations 1 month apart to establish her own intra-rater reliability and inter-rater reliability with Pluta. Reliability will be reported using the intraclass correlation coefficient, Dice similarity coefficient (a relative overlap measure) and boundary distance measures. The intra- and inter-rater reliability will be valuable data for answering hypotheses about the ability of non-expert gamers to replicate the work of anatomists.
Aim 2: To scale up labeling efforts, we will break the labeling problem into smaller tasks within a computer game.
We will break BigBrain data into small pieces to turn labeling into a parallel, distributed effort. Labeling each piece would constitute an element of a game played by non-experts.
Seed boundaries:
The expertly drawn hippocampal subfield label boundaries will only cover a small portion (2%) of the BigBrain’s hippocampus (and none of the rest of the BigBrain). We can supplement these with less accurate, automated label boundaries to get a rough sense where we should expect these boundaries to lie, by affine registering other labeled image volumes to a downsampled version of the BigBrain. We anticipate that the inter-subject anatomical variability (
Image tiles:
We will first present the labeling problem to players much as it is presented to expert labelers of neuroanatomy: as 2-D images upon which cytoarchitectonic boundaries are to be drawn (
Example boundary estimates in the BigBrain hippocampus.
This figure shows example color line drawings atop a (low-resolution) image tile containing a small portion of the BigBrain’s right hippocampus. The lines represent a novice’s estimates of the CA1/subiculum cytoarchitectonic boundary. This boundary is extremely difficult, as it results in the greatest overall disagreement among hippocampal subfield labeling protocols (
Scoring players:
Players would draw a line or curve on an image tile, and would be scored in the following ways:
Accuracy: similarity to an expert’s drawing
Intra-rater reliability: similarity to previous drawings by the player on the same tile
Inter-rater reliability: similarity to other players’ drawings on the same tile
Registration-based reliability: similarity to the seed boundaries used to select the tile
Independent reward: a score that is independent of other lines
For 1-4 above, similarity between two drawings can be computed by the Hausdorff distance measure, or similarity between regions delineated by the drawings can be measured by various overlap measures (
Training:
A new player will have no idea how to draw a boundary between cytoarchitectonic regions, and will rely on the game for initial training and assessment. A proven technique in these circumstances is to present players with “reference” puzzles where the solution is already known (see
Game mechanics:
There are a number of possibilities for the central game mechanic. One promising idea is to represent the labeling process as a drawing or painting game. This approach could appeal to the artistic side of players, especially if the game supports touch controls on smartphones and tablets. Another idea is to approach the labeling as a game about cutting or slicing, which is both a popular mechanic among mobile games and may appeal to players as being akin to brain surgery. The “surgery” would introduce a goal that is independent of labeling: the player would find a path to reach a tumor that must be removed. If the path deviates too much from the “correct” path, the player would be penalized, as in the popular “Operation” game currently manufactured by Hasbro, whereas if the path is close, the surgery is successful and another patient is presented to the player. A “correct” path could reflect one of four scores above: accuracy, intra-rater reliability, inter-rater reliability, or registration-based reliability.
With any (or all) of the types of scores, it would be possible to set up the game as a competition among players, and to display a leaderboard with players’ highest scores. Levels could be achieved based on labeling difficulty, determined by intra-rater and inter-rater reliability statistics obtained for expert labelers or for all players. Alternatively, the game could reward cooperation among players. For example, players could try to guess where others have drawn their lines on a given image tile. This would promote inter-rater reliability and therefore more consistent labels. FoldIt and EyeWire have demonstrated the power of social tools within citizen science games. In addition to a cooperative mechanic, we will consider bringing social interaction to our game via chat, team play, or badges.
The power of crowdsourcing lies not just in the number of minds focused on a challenge, but in the different and unexpected solutions that can arise. Rather than assume that the original image data will be conducive to drawing boundaries, if we were to make available browser-based image filters to enhance contrast, alter colormaps, etc., players could explore different ways of seeing and segmenting cytoarchitectonic regions. The Cancer Digital Slide Archive (CDSA) tool (below) for browser-based visualization of high-resolution image data such as the BigBrain has image filtering and semi-automated segmentation capabilities. We will explore different ways of enabling players to filter the image tiles and evaluate whether image filtering improves labeling accuracy.
Both the drawing and slicing mechanics above offer players the enjoyment of recognizing patterns and the sense of completion that comes with finishing a puzzle. These mechanics could be embellished by adding secondary mechanics such as time limits, treasures to discover, territory to surround and conquer, enemies to fight off, etc. For better or for worse, these “flashier” mechanics turn the focus away from the central scientific task, which may help to keep some players engaged and may turn others away who were attracted by the appeal of a game that purports to contribute to science and society. For this reason, we will need to strike a balance to discover which game mechanics best motivate players to contribute to the game.
Game development:
We will begin our game design process by prototyping different game mechanics (above) and testing them directly with players in order to evaluate which ideas provoke the most engagement and provide the best results. Building on this initial insight, we will use an agile software development approach to develop a polished game that we can test with larger and larger user groups. This will resemble the iterative process of design adopted by the FoldIt developers (
The ubiquity of the Web browser makes it the platform of choice for a citizen science game that wants to lower the barrier of entry to new users. We will build our game upon JavaScript and HTML5 technologies, ideally without additional plugins that require installation. This approach will enable us to draw upon Dr. David Gutman's CDSA image viewer (example viewing BigBrain data: http://node15.cci.emory.edu/BIGBRAIN/), Montreal Neurological Institute collaborator Dr. Alan Evans’ BrainBrowser, Dr. Roberto Toro's Seadragon-based drawing tool, and/or the RedWire game engine, as needed.
Data handling:
Each image tile would have a unique identifier, as would each player, and each player’s drawing. This information would be stored in a CouchDB or PostgreSQL database management system. As players play the game, image tiles would be loaded into the game (as described above), and players’ vector-based drawings would be stored in the database as JSON objects. Regarding player interaction with the game, we will link the game to the RedMetrics open game analytics service, so that such data will be accessible to citizens, scientists, and game developers alike for analysis.
Aim 3: Aggregate the results of game play to label the BigBrain.
As described above, the proposed game will result in multiple manually drawn curves for each image tile, each one corresponding to a putative cytoarchitectonic boundary for a small portion of a given histological section containing hippocampus. Aggregation of the large collection of manually drawn curves will involve the following steps:
Position the multitude of curves in the original space of the BigBrain.
Weight the confidence in each curve based on the evaluated skill or estimated performance of each player at the time it was drawn.
Propagate label boundary assignments from expert-labeled curves to all other curves.
Create a 3-dimensional, probabilistic label assignment for each volume element (voxel) based on its position with respect to all label boundaries in 3-D.
Create an additional atlas with hard boundaries based on label fusion (
Fig. ,6 Wang and Yushkevich 2013 Landman et al. 2010
Example joint fusion of multiple label assignments.
This macroscopic brain atlas was built from 20 individually labeled atlases, using joint fusion (
Exploratory Aim 1: Assess the potential for extending the game to all brain regions.
Ultimately we want to label the entire brain, so we will explore how our approach generalizes to other brain regions. Since we have expert manual labels drawn by Dr. Evans' team for some limbic regions, we will start with these as gold standard data within the game. If there is time, we will evaluate how similar players are at delineating other regions for which we do not have expert labels. For example, we can test to see how consistently players assign boundaries for cortical regions throughout the BigBrain, and see how these boundaries compare with expected number and position of Brodmann label boundaries (
Exploratory Aim 2: Train a supervised learning algorithm on the crowdsourced results. To explore the potential for learning from crowdsourced human results to automate labeling, we will train a supervised learning algorithm. We will use supervised machine learning methods in the open source scikit-learn Python package, the open source Dato Python package, and our own in the R programming environment. We have considerable experience building and training classifiers and regression models, and are very interested to try deep learning (convolutional neural network) approaches using the Dato or Theano Python packages. We have previously applied deep learning using Theano to automatically segment human brain MRI data at a macroscopic scale, and are in communication with developers of Dato for guidance in applying deep learning to the BigBrain’s microscopic data.
Contingency plans and timeline:
Timeline.
We will prepare the gold standard evaluation data (Aim 1) and develop and update the game (Aim 2) through the 18th month as we get feedback on its use. Year 2 will consist primarily of testing the aggregation of segmented data (Aim 3), exploring how well the game can generalize to segmentation of every region of the BigBrain (Exploratory Aim 1), and training and testing an automated approach that learns from the crowdsourced data (Exploratory Aim 2), as well as to publish and present our findings.
Without training or initialization, it is clear in
If players are unable to draw consistent boundaries, we will increase their training against gold-standard evaluation data and provide hints from better players and from co-registered label boundaries. If this doesn’t work, we can simplify the task and ask players if there is, or is not, a boundary within a given tile. A game that can generate a consistent set of boundary-containing tiles will be a success, because we will be able to infer the probabilistic positions of label boundaries, which may be sufficient for most label-based applications, or could be used to help guide expert labelers so that they are much more efficient.
If we fail to attract many players, we will present the game through Amazon’s Mechanical Turk (AMT). AMT provides an online, on-demand, scalable workforce, where each “human intelligence task” (HIT) is submitted by a Requester and is performed by one of tens of thousands of paid Workers around the world. AMT has been used in many scientific experiments, including cognitive behavioral experiments (
Vertebrate animals
N/A
Project
The principal aim of this proposal is to crowdsource the segmentation of brain histological data, specifically the cytoarchitectonic regions of the hippocampi of the human brain, by means of a computer game. Currently, only human experts perform reliable cytoarchitectonic labeling at a very slow and small scale, whereas we propose to enlist many human non-experts to engage in a distributed version of this task at a quick and large scale. By turning this arduous, isolated task into an engaging, collective activity, we hope to radically change the way anatomists approach segmentation/labeling.
To support this aim, we must (1) prepare expert (gold standard) labels to a subset of the hippocampal sections to evaluate crowdsourced results, and (2) aggregate the crowdsourced results to label the hippocampi. For our first exploratory aim, will explore how our approach generalizes to all other brain regions, and for our second exploratory aim, we will train a supervised learning algorithm on the crowdsourced results and evaluate how closely the automated approach matches human assessments.
We will use the BigBrain, a high-resolution 3-D model of a human brain at nearly cellular resolution (20μm isotropic) based on reconstruction of 7,404 histological sections stained for cell bodies. The BigBrain is extremely important to the neuroscience community because it represents whole-brain histological data with accompanying magnetic resonance imaging data. It is the best brain imaging data set in the world to establish a canonical space at both microscopic and macroscopic resolutions. However, for the cell-stained microstructural data to be truly useful, it needs to be segmented into cytoarchitectonic regions, which makes it the perfect focus for this project. Labeling of the BigBrain will establish an ex vivo atlas, a common space for neuroimaging data whose labels will provide a consistent, convenient, and meaningful way to communicate, classify, and analyze biomedical research data set in that space.
Call
Big Data to Knowledge (BD2K) Advancing Biomedical Science Using Crowdsourcing and Interactive Digital Media (UH2)
Hosting institution
Sage Bionetworks
Ethics and security
This proposal makes use of publicly available data.
Author contributions
AK conceived of and wrote this proposal.
References
-
Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI.NeuroImage84:505‑523. DOI: 10.1016/j.neuroimage.2013.08.067
-
Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps.Anatomy and Embryology210:343‑352. DOI: 10.1007/s00429-005-0025-5
-
BigBrain: An Ultrahigh-Resolution 3D Human Brain Model.Science340(6139):1472‑1475. DOI: 10.1126/science.1235381
-
Small neuronal size in hippocampal subfields that mediate cortical-hippocampal interactions.Biological Psychiatry35(9):717. DOI: 10.1016/0006-3223(94)91023-5
-
Pattern Separation in the Human Hippocampal CA3 and Dentate Gyrus.Science319(5870):1640‑1642. DOI: 10.1126/science.1152882
-
Staging of alzheimer's disease-related neurofibrillary changes.Neurobiology of Aging16(3):271‑278. DOI: 10.1016/0197-4580(95)00021-6
-
Vergleichende Lokalisationslehre der Grosshirnrinde.Johann Ambrosius Bart,Leipzig,?pp.
-
CogSci2011 workshop: "How to use Mechanical Turk for Cognitive Science Research".?pp. URL: http://cognitivesciencesociety.org/uploads/2011-t4.pdf
-
The challenge of designing scientific discovery games.Proceedings of the Fifth International Conference on the Foundations of Digital Games - FDG '10?:40‑47. DOI: 10.1145/1822348.1822354
-
A Retrospective Analysis of Hippocampal Pathology in Human Temporal Lobe Epilepsy: Evidence for Distinctive Patient Subcategories.Epilepsia44(8):1131‑1131. DOI: 10.1046/j.1528-1157.2003.00448.x
-
Organization and detailed parcellation of human hippocampal head and body regions based on a combined analysis of Cyto- and chemoarchitecture.Journal of Comparative Neurology523(15):2233‑2253. DOI: 10.1002/cne.23786
-
The Human Brain: Surface, Three-Dimensional Sectional Anatomy with MRI, and Blood Supply.2nd.Springer Wien,New York,?pp.
-
Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.Nature Biotechnology30(2):190‑192. DOI: 10.1038/nbt.2109
-
Crystal structure of a monomeric retroviral protease solved by protein folding game players.Nature Structural & Molecular Biology18(10):1175‑1177. DOI: 10.1038/nsmb.2119
-
Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration.NeuroImage46(3):786‑802. DOI: 10.1016/j.neuroimage.2008.12.037
-
Simultaneous truth and performance level estimation with incomplete, over-complete, and ancillary data.P roceedings of SPIE--the International Society for Optical Engineering?:?. DOI: 10.1117/12.844182
-
BigBrain: Initial Tissue Classification and Surface Extraction.2 0th Annual Meeting of the Organization for Human Brain Mapping (Hamburg, Germany)1:1.
-
Stress, Depression and Hippocampal Apoptosis.CNS & Neurological Disorders - Drug Targets5(5):531‑546. DOI: 10.2174/187152706778559273
-
Online Gamers Achieve First Crowd-Sourced Redesign of Protein.Scientific American?:?.
-
A Guide to Behavioral Experiments on Mechanical Turk.Behavior Research Methods44(1):1‑23. DOI: 10.3758/s13428-011-0124-6
-
The cognitive neuroscience of remote episodic, semantic and spatial memory.Current Opinion in Neurobiology16(2):179‑190. DOI: 10.1016/j.conb.2006.03.013
-
CBRAIN: a web-based, distributed computing platform for collaborative neuroimaging research.Frontiers in Neuroinformatics8:1. DOI: 10.3389/fninf.2014.00054
-
A pathophysiological framework of hippocampal dysfunction in ageing and disease.Nature Reviews Neuroscience12(10):585‑601. DOI: 10.1038/nrn3085
-
Multi-atlas segmentation with joint label fusion and corrective learning—an open source implementation.Frontiers in Neuroinformatics7:1. DOI: 10.3389/fninf.2013.00027
-
Hippocampal neurons in pre-clinical Alzheimer’s disease.Neurobiology of Aging25(9):1205‑1212. DOI: 10.1016/j.neurobiolaging.2003.12.005
-
A medial temporal lobe division of labor: Insights from memory in aging and early Alzheimer disease.Hippocampus21(5):461‑466. DOI: 10.1002/hipo.20779
-
Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol.NeuroImage111:526‑541. DOI: 10.1016/j.neuroimage.2015.01.004