|
Research Ideas and Outcomes : NIH Grant Proposal
|
|
Corresponding author: Arno Klein (arno@binarybottle.com)
Received: 11 Apr 2016 | Published: 13 Apr 2016
© 2016 Arno Klein, Steven P. Ellis.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Klein A, Ellis S (2016) Concurrence Topology: Finding High-Order Dependence in Neuropsychiatric Data. Research Ideas and Outcomes 2: e8815. doi: 10.3897/rio.2.e8815
|

|
Executive summary
The proposed research develops new computational tools to identify, diagnose, and predict treatment response for different mental illnesses. The research will first be applied to publicly available resting state fMRI BOLD data from patients with attentiondeficit hyperactivity disorder and autism. It will also be applied to existing clinical and biological data concerning suicidality in the context of major depressive disorder. These disorders affect millions of Americans, but these tools can be applied to any mental illness, such as Alzheimer’s disease, bipolar disorder, schizophrenia – indeed to analyze differences in brain, clinical, and biological data between any two populations.
Keywords
concurrence topology, homology, brain imaging, adhd, high-order dependence, statistical analysis
Research & Related Other Project Information
Project Decsription
There is a serious need for biomarkers to help understand, identify, diagnose and tailor treatment of psychiatric illnesses. Biomarkers could involve patterns of statistical dependence among many variables, but describing high-order dependence in many variables is a significant challenge. We propose to further develop and apply a new method, “concurrence topology”, to large psychiatric data sets with the aim of identifying biomarkers related to diagnosis, symptom clusters across diagnoses and treatment response rates by pursuing the following aims.
- To identify biomarkers related to diagnosis we will use concurrence topology for studying high-order functional connectivity in large, existing resting-state fMRI data sets.
- To aid in classification models when there are more than just a few variables, we will develop and apply concurrence topology to identify interactions, especially high-order interactions in related to diagnosis or to components of psychopathology shared across diagnostic boundaries like stress response or aggressive impulsive traits, in existing psychiatric clinical, structural MRI, and other biological data available in the PIs' institutions. These interactions might be useful biomarkers for disease.
- Concurrence topology can be very demanding computationally. To extend its reach as a tool for studying high-order dependence, concurrently with work on Aims 1 and 2 and in support of those Aims, we will rewrite our concurrence topology software to greatly increase its computing speed. We will also make it easier to maintain and easier to use by the general scientific community.
Facilities & Other Resources
Stony Brook University (SBU) and Stony Brook University Medical Center (SBUMC)
The integration of three institutions - Stony Brook University Medical Center, Stony Brook University and Stony Brook Research Foundation - provides the Department of Psychiatry and Behavioral Science with ample educational and institutional resources. These resources reside either at the medical campus, where Dr. Klein is located, or on SBU’s main campus, accessible by a 10-minute walk. Dr. Klein engages in collaborative research with faculty in the Computer Science Department and holds weekly meetings in the Computer Science building.
Within the Health Sciences Center and the surrounding campus, research is being conducted in many areas of psychiatry, neurology, and biology, allowing Dr. Klein to collaborate and consult with researchers with a broad scope of expertise. The targeted symposia and Grand Rounds presented in Health Sciences Center will expose Dr. Klein to a diverse array of outstanding psychiatric research.
The Department of Psychiatry and Behavioral Science is ideally situated to take advantage of this wealth of combined resources for training and career development of junior faculty. In addition to the department’s affiliation to Stony Brook University Hospital, there are affiliated clinical and teaching programs and Eastern Long Island Hospital in Greenport, the Northport Veteran's Hospital, and Nassau University Hospital, providing further opportunities for research collaboration. The Department also has a long history of securing external funding. There are many active research projects funded by NIH, NIMH, NIDA, NIAAA, and NCI. In addition, the department has multiple pharmaceutical industry sponsored clinical trials.
As a member of the Department of Psychiatry, Dr. Klein has access to all journal clubs and targeted symposia, in addition to the mentorship and resources of Dr. Parsey, the Chair of the Department of Psychiatry and Behavioral Science, who has been a collaborator with Dr. Klein on previous NIH-funded grants. The Department of Psychiatry and Behavioral Science provides a number of high quality clinical programs, a psychiatry residency program and accredited fellowships in child and geriatric psychiatry and an array of sponsored research activities. At present there are approximately 45 full-time faculty and numerous voluntary faculty who participate in supervision and training of students. The department also provides administrative and clerical support.
Computing resources
It is important that Dr. Klein has adequate access to a powerful computer cluster to run distributed software processes, including preprocessing workflows, Mindboggle software, and the (currently) computationally intensive concurrence topology software on thousands of brain images. This is provided in the form of a 128- core cluster that Dr. Klein maintains at Stony Brook University. In addition, both the Psychiatry and Behavioral Science Department and the imaging group have dedicated IT professionals, located on the same floor as Dr. Klein, in case computer support is needed.
Office
Dr. Klein’s office is located on the 10th Floor of the Health Sciences Center on the Stony Brook University Medical Campus. This building is connected to the Stony Brook University Hospital and within walking distance of the Stony Brook main campus.
SUBCONTRACT RESOURCES
Columbia University and Columbia University Medical Center (CUMC)
Columbia University was founded in 1754 by the royal grant of George II of England and its purpose was for the instruction of youth in the Learned Languages, and the Liberal Arts and Sciences. A medical faculty was organized in 1767, and was the first institution in the North American Colonies to bestow the degree of Doctor of Medicine. In 1814, Columbia College merged with the College of Physicians and Surgeons, which had obtained an independent charter in 1807. Its use was primarily for servicing the Presbyterian populace. However, it soon began welcoming everyone and became, as noted on a tablet that remains on the hospital today, "For the Poor of New York without Regard to Race, Creed, or Color." An agreement was signed in 1911, between Columbia University the College of Physicians and Surgeons and Presbyterian Hospital. Finally in 1928, the Columbia-Presbyterian Medical Center opened up for operation in the Washington Heights section of Manhattan.
Since that time, Columbia University Medical Center has experienced phenomenal growth and development and is now situated on a 20 acre campus and makes up almost half of the close to $2 billion budget of Columbia University. Columbia University Medical Center provides international leadership in basic, preclinical, and clinical research, in medical and health sciences education, and in patient care. The medical center trains future leaders and includes the dedicated work of many physicians, scientists, public health professionals, dentists, and nurses at the College of Physicians and Surgeons, the Mailman School of Public Health, the College of Dental Medicine, the School of Nursing, the biomedical departments of the Graduate School of Arts and Sciences, and allied research centers and institutions. Columbia University Medical Center is home to Columbia’s College of Physicians & Surgeons, which is among the most selective medical schools in the country, and the largest medical research enterprise in New York and one of the largest in the United States. It currently boasts some 13 Howard Hughes Medical Institute investigators, 47 members of the Institute of Medicine, 26 members of the American Academy of Arts and Sciences, and 16 Nobel laureates in Medicine or Physiology – two of which are presently on staff. For more information, please visit www.cumc.columbia.edu.
The New York State Psychiatric Institute (NYSPI)
Formerly the Pathological Institute, the New York State Psychiatric Institute was founded in 1896 and began its affiliation with the Columbia University College of Physicians & Surgeons in 1925. It has since then grown into a pioneering world-renowned facility for the advancements in mental health research and hygiene. The New York State Psychiatric Institute (NYSPI) has as its mission “the search for knowledge about the causes, prevention, and treatment of mental illness.” It is a vibrant department and is chaired by Dr. Jeffrey Lieberman, an international expert in the area of schizophrenia treatment.
Laboratories are located in the north part of the building and space for inpatient care, outpatient clinics, and education is in the south part of the building. Two enclosed bridges connect the new building to the Kolb Annex and the Milstein Hospital Building. In 1985 the Lawrence C. Kolb Research Annex was added to the original facility, which has 50,000 square feet of laboratory space plus the clinical research units described above, office library, and other areas designed for its functioning as a hospital. The Annex also contains the Howard Hughes Institute supporting Nobel Laureate Dr. Eric Kandel and affiliated investigators. This state supported facility is on the campus of the CUMC and has been one of the leading institutions for psychiatric research for over 100 years. Its resources, research faculty and staff, combined with those of the Research Foundation for Mental Hygiene at NYSPI and Columbia University Department of Psychiatry, have made it one of the nation’s foremost psychiatric research centers.
Clinical, laboratory, and support facilities
The New York State Psychiatric Institute’s newest building was dedicated in May of 1997 and is now known as the Pardes Building. This 6-story building contains approximately 330,000 sq ft of state-of-the-art laboratory, clinical research and educational facilities, including the inpatient research units and outpatient clinics that are heavily utilized by the fellows and mentors in this program. Trainees also have access to a host of laboratories and clinical facilities at CUMC and NYPH, especially the structural and functional imaging resources in the Department of Radiology, the Irving Center for Clinical Research (GCRC) and the Center for Neurobiology and Behavior, which are all in nearby buildings. Trainees have full access to an excellent library, computer facilities, photocopying, and the animal care facilities in the Kolb Annex building and elsewhere at CUMC.
The Department of Psychiatry/Division of Molecular Imaging and Neuropathology (MIND) at Columbia University/NYSPI comprises several components: wet laboratories including biochemistry, neurophysiology, molecular biology, pharmacology, neurohistology and an image analysis facility for quantitative autoradiography and quantitative morphometry. The MIND division has clinical office space for screening, evaluation, and treatment of study participants. We also have a bank of -80°C freezers for storage of samples. The MIND Computer Center is equipped with several multi-core Apple PowerMac and MacPro workstations running Mac OS X 10.5 and 10.6. The lab is networked via 1000Mbps (gigabit) copper Ethernet and CISCO Catalyst switches. The workstations connect to a Dell Poweredge 6600 computational server with four Intel Xeon MP 2.50GHz CPUs and 24GB of RAM running RedHat Enterprise Linux 3. This server also provides a collection of web applications built using a MySQL database, the lighttpd web server and the Django framework. The workstations also connect to a multiprocessing computational cluster, comprised of 48 Apple XServe G5 nodes each with 2 2.3GHz CPUs and 4GB RAM, running Mac OS 10.4 and the Matlab Distributed Computing Engine as the job manager. The head node of the cluster also provides NTP and DNS services to both the workstations and the other servers. A subset of the workstations also forms a second small multiprocessing cluster, running both the Matlab Distributed Computing Engine and the Sun Grid Engine depending on the needs. The primary storage is formed by a Sun Fire X4500, containing 48 500-Gb SATA drives and using the ZFS filesystem for a total storage space of 20TB. This storage area is shared over NFS. A second, smaller file server is formed by an Apple XServe G5 containing two mirrored SATA drives. This server shares files over SMB, AFP and HTTP. The whole storage area is backed up over the network by an independent Linux box and stored on a linear SATA disk array. Printers include a Samsung color laser printer and several high capacity HP black and white printers. The facility also hosts 2 40U cabinets powered with 4 3phase 20Amp circuits to which they are backed up by a generator.
Office
Dr. Ellis, the Co-Principal Investigator, has an office on the second floor of the New York State Psychiatric Institute on the Columbia University Medical Campus. He programs in R on a MacIntosh Pro desktop computer.
Specific research plan
Specific aims
A major research priority in medicine and psychiatry is the search for biomarkers to diagnose neuropsychiatric illnesses, characterize symptom clusters across diagnoses and predict treatment response (Insel and Cuthbert, 2009). Understanding interactions involving variables such as age, gender, clinical status, drug tolerance, and treatment, i.e., biomarkers involving patterns of statistical dependence among multiple variables, can be critical for patients’ outcome and survival. For k=2,3,..., define “kth-order dependence” to be that which can be discerned in groups of k variables, but no fewer. Principal components analysis and many other standard multivariate statistical procedures only describe dependence of order 2. But dependence of order greater than 2 (“high-order” dependence) can be important.
Statistical dependence can be studied with regression-type or structural equation modeling, but the biological relationships that underlie these models are not always understood. Another approach is to conduct “agnostic” analyses that make few assumptions and treat all variables the same a priori. Regression analysis, for example, is not agnostic because it requires designating some variables as responses and others as predictors. A challenge for extracting meaningful biomarkers is to agnostically describe high-order dependence among many variables in an interpretable fashion.
We have developed a new statistical method, "concurrence topology", that can be used to succinctly describe high-order dependence in dozens of variables in an agnostic fashion. Concurrence topology represents multivariate data as a series of abstract shapes and describes the topology of those shapes. We have applied it in a preliminary analysis (article under review) to find differences in high-order functional connectivity in samples of 25 attention deficit hyperactivity disorder (ADHD) subjects and 41 controls. Using concurrence topology, we found numerous differences in high-order dependence structure in the two groups, including a robust difference in 6th-order dependence in individual subject’s fMRI data, and even evidence of a difference in 7th-order dependence. This feat is difficult, if not impossible, with other statistical methods because a naive, but agnostic, approach would likely attempt to summarize all 3,365,856 different groups of seven brain regions among 32 regions. Instead of looking at the 3,365,856 trees, concurrence topology finds patterns in the forest. But for limits in computing speed, we could have investigated even higher orders of dependence (Aim 3).
We are proposing to use concurrence topology to obtain descriptions of high-order dependence to help identify novel biomarkers, by pursuing the following aims:
- Aim 1. To uncover high-order functional connectivity biomarkers in individual subject data, we will first confirm and extend our brain imaging findings in the much larger ADHD-200 fMRI data set (776 subjects), and second, apply our method to the ABIDE autism spectrum disorder (ASD) resting-state fMRI data set (1,110 subjects). We will use concurrence topology to classify individuals by diagnosis and even subtype. The large sample sizes will allow cross-validation to reduce the false positive rate.
- Aim 2. To find biomarkers involving non-time series and non-imaging data based on group-level patterns of high-order dependence, we will first refine our method for doing this and then apply it to dozens of variables from different domains. These variables include clinical, demographic, behavioral, questionnaire, genotype, etc. in pre-existing data to look for diagnostic biomarkers for ADHD, ASD, suicidal behavior, and treatment response in major depressive disorder. Again, large sample sizes will permit cross-validation.
- Aim 3. To apply concurrence topology with more variables (regions, in fMRI applications), more fMRI subjects, and to examine higher levels of dependence, we will, concurrently with and in support of Aims 1 and 2, rewrite our software to make it faster and easier to read and maintain for wider adoption by the scientific community.
The above Aims permit us to further develop and apply our new statistical approach to extract biomarkers as patterns of high-order dependence in neuropsychiatric data. Our long-term goal is to uncover the biological relationships that underlie the patterns of dependence we find with concurrence topology to improve our understanding of the pathophysiology in these and other neuropsychiatric illnesses.
Research strategy
1. Significance
1.1. Meeting the challenge of biomarker discovery
Diagnosis of mental disorders suffers from a dearth of reliable biomarkers [
ADHD "affects at least 5-10% of school-age children and is associated with substantial lifelong impairment, with annual direct costs exceeding $36 billion/year in the US. Despite a voluminous empirical literature, the scientific community remains without a comprehensive model of the pathophysiology of ADHD. Further, the clinical community remains without objective biological tools capable of informing the diagnosis of ADHD for an individual or guiding clinicians in their decision-making regarding treatment." (http://fcon_1000.projects.nitrc.org/indi/adhd200/)
ASD "are now recognized to occur in more than 1% of children, causing immense suffering to individuals and their families." (http://fcon_1000.projects.nitrc.org/indi/abide/)
MDD has an overwhelming impact on the health of Americans, as noted by the NIMH (http://www.nimh.nih.gov/health/publications/the-numbers-count-mental-disorders-in-america/index.shtml). It is the leading cause of disability in the U.S. for ages 15-44 and affects approximately 14.8 million American adults, or about 6.7 percent of the U.S. population age 18 and older in a given year (http://www.who.int/healthinfo/global_burden_disease/GBD_report_2004update_AnnexA.pdf, http://www.census.gov/popest/national/asrh/).
Suicide is the eleventh leading cause of death in the United States with over 30,000 individuals committing suicide per year [
We attribute the elusiveness of biomarkers partly to the fact that traditional methods used to analyze neuropsychiatric data do not adequately reflect their complexity. The Research Domain Criteria project (RDoC) of the NIMH encourages investigation of “functioning ... across multiple units of analysis, from genes to neural circuits to behaviors, cutting across disorders as traditionally defined.” This is facilitated by statistical analysis of variables across several domains at once. This cannot be done with just a handful of variables.
In the proposed research we will further develop and apply a new statistical method to capture this complexity in functional connectivity data [
1.2. Taking advantage of large data sources for extracting biomarkers
Advances in neuroimaging brain activity have opened up tremendous stores of rich data from which biomarkers may be drawn (see sections 3.5.1 and 3.5.2 for descriptions of the data we will use). An important aspect of brain activity is the interaction among brain regions. This interaction is reflected in “functional connectivity” in neuroimaging time series, which is “statistical dependencies between spatially segregated neuronal events” [
Functional neuroimaging data alone tell only a part of the story about the health of an individual. Phenotypic data (and non-time series imaging data) of many kinds are routinely acquired from patients and should be included in a search for biomarkers of complex neuropsychiatric illnesses. Aim 2, consistent with RDoC, is concerned with describing or summarizing data from multiple sources (3.5.2), not simply functional connectivity times series brain imaging data. (However, summaries from analyses performed under Aim 1 can serve as inputs to the sorts of analyses of Aim 2.) The method with which Aim 2 is concerned starts with group-level statistics and translates them into the level of the individual.
Our longterm goal is to use these functional connectivity and multiple-source biomarkers to gain insight into the pathophysiology of mental illnesses for prediction of disease course and treatment response in individual patients to better personalize treatment accordingly.
1.3. Overcoming problems with current approaches for extracting biomarkers
Individual variables might serve as biomarkers. Many statistical methods can help identify such variables. But patterns of the joint distribution of variables (i.e., the form of statistical dependence) might also serve as biomarkers. One way to study these patterns is to specify a regression, classifier, or structural equation model involving only a few variables. (We lump these methods together under the generic term “model”.)
Dependence structure often has an “order”: If a feature of the joint distribution of variables can be detected by looking at k variables at a time, but not by looking only at k−1 variables at a time, then that feature reflects “kth-order dependence” among the variables. For example, traditional cluster analysis of variables, (Pearson, Kendall, and Spearman) correlation, factor analysis (including principal components analysis), and linear discriminant analysis are measures of 2nd-order dependence because those analyses can all be carried out by looking at the variables two at a time. “High-order” dependence is dependence of order at least three. A model including interactions registers high-order dependence.
However, one might not wish to posit such a specific model, but instead proceed “agnostically”. Data analysis is “agnostic” if, a priori, all variables are treated the same (for k = 1, 2, 3,... all groups of k variables are treated the same) and few a priori assumptions are made about the nature of the joint distribution. The aforementioned cluster, correlation, etc. analysis methods are all agnostic.
If there are many variables it is difficult to capture high-order dependence agnostically using a model. In general, in agnostically analyzing increasingly high orders of dependence in large numbers of variables one must overcome a “combinatorial explosion”. Agnostically studying kth-order dependence means looking for a pattern in all “k-tuples” of variables, i.e., all groups of k variables, of which there can be very many. In our preliminary fMRI analyses (3.4.1) we examined 2nd- (2,701 pairs), 3rd- (64,824 triplets), and 4th-order dependence (1,150,626 quadruplets) in 74 variables. We also looked at 7th-order dependence in 32 variables (Example 1). That analysis involves 3,365,856 septuplets. So on the face of it if one wished to study 7th-order dependence among 32 variables one would have to make sense of 3,365,856 numbers.
Statistical methods such as independent components analysis [
1.4. Concurrence topology
In this proposal, we discuss a new method, “concurrence topology”, that makes remarkable headway in overcoming the combinatorial explosion without the drawbacks of the above-mentioned statistical methods. The paper by Ellis and Klein [
Concurrence topology is used on dichotomized data. It works by describing kth-order dependence, not directly in terms of k-tuples, but in terms of larger structures called “homology classes”. Concurrence topology begins by representing multivariate data as a series of abstract shapes. Homology classes are just holes in the shapes and represent what might be thought of as inhibitory relationships. Cluster analysis can be used to study the dependence among variables. Viewing cluster analysis as a method for finding, not clusters, but gaps between clusters, concurrence topology can be thought of as “cluster analysis on steroids.” One advantage of concurrence topology is that it radically reduces the size of the selection problem from selecting from thousands or millions of k-tuples to selecting from a few dozen (usually fewer) homology classes. This gives one a reasonable chance at managing the multiple comparisons problem.
Concurrence topology can reveal complex relationships within dozens (but not 100s or 1,000s) of variables. Hence, it can draw connections among multiple domains (e.g., genes, circuits, behavior). Another advantage is that, in principle, concurrence topology can be used to investigate dependence of any order. (The obstacle is computational, addressed in Aim 3.) Concurrence topology is also explicit: homology classes are associated with specific orders of dependence and can be “localized” (3.3) to reveal specific groups of variables most closely associated with them. Since concurrence topology can be computed from time series data from just one subject (Aim 1), summaries of patient concurrence topology could potentially be used for diagnostic purposes. In the sort of analyses proposed under Aim 2, group-level descriptions of dependence are translated into individual-level interaction variables that can again be used for diagnosis.
2. Innovation
This proposal is innovative with regard to the methods we are developing, the domain to which we are applying them, and to the results we are seeking.
Methods: Concurrence topology is a new approach to multivariate data analysis that applies algebraic topology to statistics and is radically different from other currently used statistical methods. Not only does it more succinctly summarize high-order dependence than most conventional agnostic statistical methods do, but concurrence topology captures very different aspects of high-order dependence and therefore complements other approaches. (But we apply conventional statistical methods to concurrence topology output; 3.4.)
Domain and results: We apply topology for the first time to resting state fMRI (functional connectivity) data and to non-imaging phenotypic neuropsychiatric variables and use topology for the first time to extract biomarkers of neuropsychiatric illnesses.
3. Approach
3.1. MRI data processing
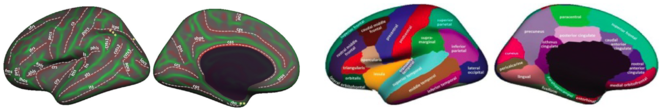
PI Dr. Klein is involved in the development of state-of-the-art software workflows for MRI data processing (article under preparation; https://github.com/INCF/BrainImagingPipelines) and is the main developer of the Mindboggle software package (http://mindboggle.info) that automates cortical labeling (
Brain labeling protocol (left, on an inflated cortex) and cortical labels (right) used by Mindboggle for extracting regions analyzed by our concurrence topology software.
3.2. Persistent homology
Concurrence topology describes high-order dependence in terms of homology classes. Selection of homology classes is based on their “persistence” [
Persistence plot showing third- and higher-order dependence in the fMRI BOLD data from an individual control subject (as in Aim 1). The larger disk indicates two coinciding points. The point with the asterisk is discussed in 3.4.1, Example 2.
3.3. Localization
Often researchers will want detailed information concerning high-order dependence in order to gain understanding of a disease process. Concurrence topology can tell more about a persistent homology class than just what is revealed in a persistence plot. A homology class involves all variables, but some variables are involved more directly than others. For purposes of interpretation (and for finding interactions, 3.4.2) it is important to find the variables directly involved in persistent classes, at least for those with long lifespans. These can be found as members of “short cycles”. A “cycle” is a structure in the data that wraps around the hole (or holes) represented by the homology class. A cycle belonging to a homology class representing kth- and higher-order dependence can involve no fewer than than k variables. Those involving only k variables are “short cycles.” Such short cycles are those most intimately associated with the corresponding hole and capture dependence of order exactly k. The process of finding short cycles is “localization”. See 3.4.1 (Example 2) and
3.4.2 for applications of localization.
3.4. Preliminary results
3.4.1. fMRI data analyses
Most of our effort in using concurrence topology has been in applying it to resting state fMRI blood oxygenated-level dependent (BOLD) data [
We applied concurrence topology to each subject’s data separately and computed summaries of the results. Then we used standard statistical methods to compare the subject-wise concurrence topology summaries between the two groups. Concurrence topology can be applied to multivariate time series either in the time or Fourier domains [
Example 1. Fewer ADHD subjects (64.0%) had any holes corresponding to dependence of order six or higher in the time domain in the DMN than did controls (92.6%). Thus, the two groups differ in their pattern of 6th-order dependence. There was a less robust finding of the same sort in 7th-order dependence (the “7th-order dependence in 32 variables” referred to in 1.3). We also found differences in the whole brain in the Fourier domain in dependence orders 3 and 4.
Example 2. The persistent class marked with an asterisk in
Being able to agnostically pinpoint specific triples of regions that discriminate the two groups would be difficult with any other statistical method.
3.4.2. Interactions in logistic regression for identifying suicide attempters
We used concurrence topology for interaction identification in Hamilton Depression Rating Scale (HDRS) data on 700 subjects randomly drawn from a clinical database in the M.I.N.D. division of the New York State Psychiatric Institute (NYSPI), where PI Dr. Ellis works. Our goal was to use the HDRS items (excluding suicide item) to discriminate subjects with and without a history of suicide attempt. We further split the 700 subjects into a “training” and “testing” sample. Using concurrence topology we found in the training sample persistent homology classes that distinguished the two groups. Examination of these led to one 3rd-order interaction and three 2nd-order interactions. We fitted lasso [
3.5. Proposed work
3.5.1. Individual functional connectivity data (Aim 1)
We will first confirm and extend the our brain imaging findings in [
We also have functional connectivity brain imaging data from patients with MDD to which we can apply our method to attempt to predict antidepressant treatment response. PI Dr. Klein is a Co- Investigator on the “Biosignature Discovery for Personalized Treatment of Depression” (1U01MH092250- 01), a large, multi-site project acquiring multimodal imaging data from 400 individuals with MDD, specifically designed to make data available to find biomarkers for MDD.
3.5.2. Non-temporal group data (Aim 2)
We will further develop our method to first detect high-order dependencies in multiple sources of nontemporal, non-imaging data at the group level and then translate them into individual level interaction variables. These interaction variables can be included in classification modes such as logistic regression as described in 3.4.2. We will apply the method to classify clinical subtypes of ADHD and of ASD by accompanying phenotypic information (symptom measures, lifetime medication status, behavioral measures, IQ scores, etc.) in the ADHD-200 and ABIDE data sets.
The M.I.N.D. division at NYSPI has large collections of clinical (over 1,500 subjects, over 4,000 variables), neuropsychological (over 500 subjects), genotype (almost 500 genotyped, over 500 with DNA gene chips), postmortem (autoradiograms of brains of over 250 subjects), and structural MRI (over 400) data pertaining to suicide and MDD. With the guidance of Co-Investigator Dr. Mann we will use clinical and/or biological criteria to select samples of subjects and up to 100 or so variables across several domains (as envisioned by RDoC) for concurrence topology analysis. The database manager (TBH) will then put together analytic data sets from the NYSPI databases.
3.5.3. Software development (Aim 3)
PI Dr. Ellis has written a package of programs in the statistical programming R language (http://www.rproject.org) for doing concurrence topology analysis. Currently, the amount of computer time needed by concurrence topology varies greatly from data set to data set and in extreme cases can require more than one week to analyze one data set. The probability of having a long computation increases the more variables there are and especially with order of dependence being analyzed. To increase the computational capabilities of our software, we propose to make the software run much faster. To do this, Dr. Ellis, assisted by the programmer (TBH), will rewrite the most computationally intensive portions of the software in a compiled language, such as C. Increasing computational speed will be one of the first undertakings of the project because of its importance for further development of the algorithms and their application to the large data sets described above. Some data structures recur many times in our software. In order to improve maintainability of the code, we will employ object-oriented programming [
PI Dr. Klein has considerable programming experience, having developed the Mindboggle software described above (3.1), and will be involved in the software engineering challenges of this proposal, ensuring that the project adopts modern practices of test-driven development and distributed version control (hosted by http://github.com). In addition to working from the current code base, he has some experience with and will further explore the capabilities of the Dionysus software, a C++ library with Python bindings for computing persistent homology developed by Dmitriy Morozov.
In addition to Dr. Morozov, Prof. Konstantin Mischaikow at Rutgers University, an expert on computational topology, will serve as an unpaid consultant for the project.
3.6. Timeline
Aim 1. We will spend the first year and a half seeing if our ADHD brain imaging findings will replicate in the much larger ADHD-200 data set with 776 subjects, and applying our method to find individual functional connectivity biomarkers in the ABIDE ASD data set with 1,110 subjects, as well as the MDD data set with 400 subjects. We will try to discriminate between different subtypes of the disease using concurrence topology, and evaluate using cross-validation.
Aim 2. We will run analyses for identifying non-time series group data biomarkers to diagnose subtypes of ADHD and ASD and predict treatment response in MDD and suicidal behavior (five months for each condition, including time to write, submit, and revise publications). In Year 2, we will include in the ADHD and ASD analyses subject-level summaries of high-order dependence computed under Aim 1.
Aim 3. We will spend the first six months making the concurrence topology software faster, and in the following year, we will restructure the code to follow an object-oriented framework that will help make the code base more concise and easier to maintain. We will then write a paper describing and publicizing the software. During this period, we will also determine which portions of the Dionysus computational homology software could be used to advance our concurrence topology software, and if so, extend the Dionysus code base to do concurrence topology in case we find it faster and more appropriate to develop with this code base.
Resource sharing plan
Multiple Project Directors/Principal Investigators (Pds/PIs) Leadership Plan
Rationale for the multiple Pis
The project proposes multiple Principal Investigators, one at Stony Brook University Medical Campus and the other at Columbia University Medical Campus, to capitalize on the specific expertise of Dr. Klein and Dr. Ellis. Because it proposes to develop computational algebraic topological and statistical methods to establish biomarkers based on region and feature extraction and processing from brain data, it is essential to have significant expertise in mathematics and statistics (Ellis) as well as expertise in brain image processing and region and feature extraction (Klein). Because of this clear division of expertise, they intend to resolve conflicts by deferring to colleagues in their respective fields. Dr. Klein will defer to the chair of his department, Dr. Parsey, and Dr. Ellis will defer to Co-Investigator Dr. J. John Mann and consultants Drs. Mischaikow and Ferry. For several years Dr. Klein and Dr. Ellis have been exploring the use of concurrence topology in brain imaging. They have written a paper on the subject which has been submitted to a statistics journal. So it is natural that they team up to perform the work described in the proposal.
Expertise of Principal Investigators
Dr. Arno Klein is a Research Assistant Professor of Psychiatry and Behavioral Science at Stony Brook University Medical Campus. Dr. Klein’s research focuses on brain imaging, image processing, and information visualization. Dr. Klein received a B.S. In Biopsychology from the University of Michigan in 1993, an M.S. In Media Arts and Sciences from M.I.T. In 1996, and a Ph.D. In Neuroscience from the Weill Medical College of Cornell University in 2004. Prior to his appointment at Columbia University, Dr. Klein worked as an imaging research analyst at Columbia University, and as an Information Synthesis Theorist and Program Analyst specializing in complex data visualization at the Parsons Institute for Information Mapping at the New School in New York. He is an expert in neuroinformatics and has published the largest registration and brain extraction algorithm evaluation studies ever conducted, and has recently led a group to create the world’s largest manually labeled data set of brain images in the world [
Dr. Steven Ellis is an Associate Professor of Clinical Neurobiology (in Psychiatry) at Columbia University. Dr. Ellis will play a major role in planning analyses of within-subject functional connectivity. His primary effort will be to (1) apply concurrence topology to existing large clinical and biological data sets available in the Molecular Imaging and Neuropathology Division in the New York State Psychiatric Institute at Columbia University to discriminate clinical populations (defined, for example, by diagnosis and/or treatment response). He will also (2) further develop the concurrence topology software. Dr. Ellis’s interests include topological aspects of multivariate data analysis. He also has a strong interest in statistical computing. He is Director, Statistics and Computing Core, Conte Center for the Neuroscience of Mental Disorders (CCNMD): The Neurobiology of Suicidal Behavior. He invented the concurrence topology method and wrote software for its implementation.
Call
unsolicited R21 (2013)
Hosting institution
Stony Brook University
Ethics and security
Only publicly available data will be used.
Author contributions
AK and SE wrote this proposal in 2013.
References
-
Structure and Interpretation of Computer Programs.MIT Press,Cambridge.
-
Latent Variable Models and Factor Analysis.Wiley Series in Probability and Statistics,277pp. URL: https://doi.org/10.1002/9781119970583 DOI: 10.1002/9781119970583
-
Time Series: Data Analysis and Theory.Classics In Applied Mathematics, Philadelphia.Society for Industrial and Applied Mathematics,Philadelphia. DOI: 10.1137/1.9780898719246
-
An introduction to tensor products with applications to multiway data analysis.Chemometrics and Intelligent Laboratory Systems28:229‑237. DOI: 10.1016/0169-7439(95)80060-m
-
Centers for Disease Control and Prevention. National Center for Injury Prevention and Control, Webbased Injury Statistics Query and Reporting System (WISQARS). www.cdc.gov/injury/wisqars/index.html
-
Cell Groups Reveal Structure of Stimulus Space.PLoS Computational Biology4(10):e1000205. DOI: 10.1371/journal.pcbi.1000205
-
Nonparametric Bayes modeling of multivariate categorical data.J. Amer. Statist. Assoc104(2009):1042‑1051. DOI: 10.1198/jasa.2009.tm08439
-
Computational Topology: An Introduction.American Mathematical Society,Providence.
-
Describing high-order statistical dependence using “concurrence topology”, with application to functional MRI brain data.Homology, Homotopy and Applications16:245‑264. URL: http://arxiv.org/abs/1212.1642
-
The Elements of Statistical Learning: Data Mining, Inference, and Prediction.Springer,New York.
-
Exploring the brain network: A review on restingstate fMRI functional connectivity.European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology20(8):519‑534.
-
Independent Component Analysis.Wiley,New York.
-
The under-recognized public health crisis of suicide. http://www.nimh.nih.gov/about/director/2010/the-under-recognized-publichealth- crisis-of-suicide.shtml
-
Endophenotypes: bridging genomic complexity and disorder heterogeneity.Biological psychiatry66(11):988‑989. DOI: 10.1016/j.biopsych.2009.10.008
-
Functional MRI: An Introduction to Methods.Oxford University Press
-
101 Labeled Brain Images and a Consistent Human Cortical Labeling Protocol.Frontiers in Brain Imaging Methods6:171. DOI: 10.3389/fnins.2012.00171
-
Mindboggle - A neuroinformatics framework for automated anatomical labeling and shape analysis of brain images.Society for Neuroscience 2011.1pp.
-
Mindboggle - An informatics framework for open research in quantifying the shape of the human brain.BioImage Informatics II 2011 Conference at Janelia Farm.1pp.
-
Automated extraction of nested sulcal features from human brain MRI data.Human Brain Mapping 2011.1pp.
-
A graph-based database of hierarchical brain features.Neuroinformatics 2011.1pp.
-
Elements of Algebraic Topology.Benjamin/Cummings,Menlo Park, CA.
-
Analyzing effective connectivity with functional magnetic resonance imaging.WIREs Cogn Sci1:446‑459. DOI: 10.1002/wcs.58
-
Functional connectivity of default mode network components: Correlation, anticorrelation, and causality.Human Brain Mapping30:625‑637. DOI: 10.1002/hbm.20531