|
Research Ideas and Outcomes : Grant Proposal
|
|
Corresponding author: Peter Murray-Rust (pm286@cam.ac.uk)
Received: 09 Mar 2016 | Published: 10 Mar 2016
© 2016 Maryann Martone, Peter Murray-Rust, Jenny Molloy, Tom Arrow, Mark MacGillivray, Chris Kittel, Stefan Kasberger, Graham Steel, Charles Oppenheim, Anusha Ranganathan, Jonathan P. Tennant, Jon Udell.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Martone M, Murray-Rust P, Molloy J, Arrow T, MacGillivray M, Kittel C, Kasberger S, Steel G, Oppenheim C, Ranganathan A, Tennant J, Udell J (2016) ContentMine/Hypothes.is Proposal. Research Ideas and Outcomes 2: e8424. doi: 10.3897/rio.2.e8424
|

|
Abstract
Everyone's talking about Zika. Should we go to Brazil? Put off having babies? What are the actual facts? Many people - not just "academics" and "medics" - want to fact-check what they hear on the news by reading the scientific literature and form their own opinions. But how should they start?
Public resources of over a million biomedical papers exist, like PubMedCentral (PMC) and Europe PubMedCentral (EPMC). The ContentMine (CM) team, funded by the Shuttleworth Foundation, has worked with EPMC to create a unique interface to this literature. Search for 'Zika' and `getpapers` downloads all the papers automatically. You don't just get the papers, but also a novel, faceted view of what’s in them and other topics people are talking about. You might be surprised, but the most common species mentioned is not the virus, but a mosquito - Yellow Fever Mosquito (Aedes aegypti), because it spreads Zika. Latin names are very helpful - there's no confusion and all scientists use them. They're one of several entities that our software can discover, including genes, DNA, places, and chemicals. These observations or mentions are ‘facts’ and importantly, facts can’t be copyrighted so we can share them with anyone.
But are they important? That's where Hypothes.is comes in. It's a way of commenting on papers and sections in them. "Have you noticed that Wolbachia (a bacterium that stops mosquitoes transmitting viruses) occurs in several papers? see the Results section here ..." , "No! that's worth following up." Hypothes.is supports communities forming around facts and knowledge that anyone can create and participate in - not just academics. We propose that ContentMine and Hypothes.is combine to creat an amanuens.is - a scholarly assistant that automates the flow of new facts to the experts and communities of interest and experts. Through combining machines and humans in a tight, iterating, loop, amanuens.is will be able to mine critically important facts and make them available to the world.
Keywords
amanuens.is, Zika, open science, open access, ContentMine
Introduction
Sixty-five years ago Joseph Licklider foresaw “Man-Computer Symbiosis” where “"[humans] will set the goals, formulate the hypotheses, determine the criteria, and perform the evaluations. Computing machines will do the routinizable work that must be done to prepare the way for insights and decisions ..."(
Akua is a postdoc at the International Development Research Association for Africa (IDRAA). She's starting work in a multisite, multidisciplinary research project to observe signs of a Zika outbreak coming to Africa and prepare a response. What is the likelihood? What are the options to prevent the disease? She needs to get up to speed quickly. She uses amanuens.is to search for ‘Zika’ in all biomedical papers.
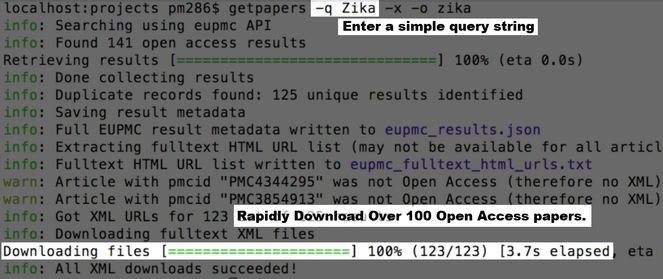
Within 3 minutes amanuens.is has downloaded all Open Access papers on Zika from Europe PubMedCentral (EPMC) and elsewhere (
Akua can quickly scan through the abstracts or papers but can also see a table of facts, visualise, search and link them (
A data table showing facts extracted from the 123 papers, including species, human genes, DNA primers and top word frequencies.
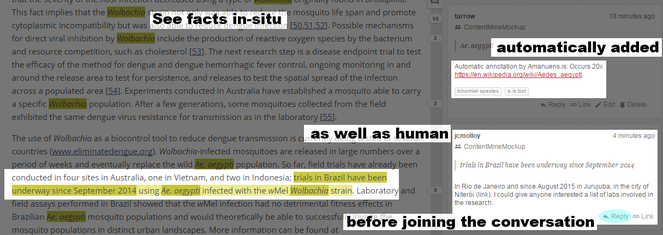
There are facts on places, on other tropical diseases. On genes. On species. The most common mosquito mentioned is Aedes aegypti. Akua knows about Ae. aegypti - it spreads Yellow Fever and Dengue and also Zika virus. That's a great start. But the most reported genus is something she's never heard about: Wolbachia. Amanuens.is pulls community-curated information from Wikipedia and Wikidata so she rapidly learns that Wolbachia is a bacterium that can stop some mosquitoes transmitting various viruses. That sounds worth exploring. But amanuens.is tells her more - there's a Hypothes.is community working on annotating the literature for Wolbachia (
Amanuens.is has automatically highlighted facts which can be viewed alongside manual annotations. Annotators can discuss and reply to each other in the browser.
Hypothes.is is a mixture of software and communities to annotate the literature. Through a web-based plug installed in their browser, anyone can layer comments tagged to specific sections of a web document, including tables. Annotations can include links, pictures or videos. When a member finds a paper of interest, they can mark up sections: "Interesting!", "Invalid - that was recently shown to be flawed [ref]", "'Funded by GillBates'. GB is really committed to combating mosquito-borne disease - I can put people in touch with their Africa office". They can insert GoogleMaps pinpointing locations mentioned or videos explaining a technique. If links within these papers are broken, users can supply correct ones.
Through Hypothes.is, Akua can reply to annotations to ask additional questions and will be notified if someone responds. In a future release of Hypothes.is, she will be notified if new annotations are added or she can put a “watch” on a paper to be notified if someone annotates it. She will also be notified if annotators anywhere mention or tag with Zika or terms. Even if the papers are annotated in another location, Hypothes.is will sync the annotations to the EPMC version.
So Akua is connected with Wolbach.is - the Hypothes.is Wolbachia community - and through using the ContentMine she has already contributed to their annotations. She invites them to write a brief proposal and they rapidly mobilise resources to test for Zika resistance in Wolbachia-infected mosquitoes. Finding facts to finding people took 15 minutes and this is how modern collaborative science should work. The people then create knowledge from the facts. The knowledge creates communities. The communities explore science- and people-based solutions.
Amanuens.is has also highlighted that there's a Zika forest in Africa, where the virus was discovered half a century ago. Akua remembers that the Ebola outbreak in Liberia had actually been predicted in a paper 30 years ago but the world didn't have amanuens.is then and so it wasn't re-discovered. Maybe some of those old papers about Zika forest have clues about the disease? If they do, amanuens.is will help find and critique them!
Why amanuens.is and who we are
Scientists and clinicians publish thousands of papers each year which are vital to direct efforts to improve health. However this ever increasing quantity of valuable, peer-reviewed information needs to be distilled so the right people the information to make the right decisions in a timely manner. Current attempts to do this, such as systematic reviews by Cochrane, are very expensive, time-consuming and tedious for the humans involved. Thankfully the data and knowledge needed to address the health crises like Ebola and Zika virus are becoming more open, but we lack tools that are both flexible and intuitive enough for biomedical researchers to manage this information overload. We believe that a combination of intelligent machines and crowdsourcing by annotation provides a solution in the form of a 21st century scholarly assistant for the digital age, much like a trusted laboratory assistant or ‘amanuensis’.
ContentMine are building an open source pipeline to extract facts from scientific documents that will make the literature review process cheaper, more rigorous, continuous and transparent. We access papers through APIs and web-scraping, normalise the documents and then extract facts such as species names, diseases, DNA sequences, chemicals and sets of keywords through a combination of pattern matching and dictionaries. This approach is highly customisable, extensible and understandable to all researchers. The outputs are suitable for a wide range of uses and users: for example, you can export already detailed tables and spreadsheets of data in addition to developer-friendly XML. We publish these facts openly online, making them available to the 99% of the population who have no access to most of the scholarly literature but might still be interested and invested in updates on, for example, disease outbreaks. These people might include the 3,500 editors of the English Wikipedia entry for Ebola, or dedicated citizen scientists who are getting interested in certain proteins after playing FoldIt. We see indiscriminate and open sharing of scientific facts as absolutely key to an inclusive open science ecosystem.
We have been impressed with the power of simple filtering of facts and how it guides us to further exploration. The fact-vectors provided by an initial query (“Zika”) prompt new queries (“Where’s Wolbachia?”). Because the software is modular it’s straightforward to automate this iteration - we can expand queries to additional organisms, but even more relevantly, to policy studies, sociology and economy of the tropics. What social factors are correlated with insecticide resistance? How have diseases spread from Africa to other continents? These are the sort of questions we would like to provide tools to explore.
Facts are important - but science is performed by people - so ContentMine are partnering with Hypothes.is to bring communities together around facts in the scholarly literature. Hypothes.is have already attracted the support of a coalition of scholarly publishers to explore annotation of journal papers and other research outputs through an open and portable annotation layer. They are actively engaging with researchers, for instance through partnering with the Neuroscience Information Framework to automatically insert information on reagents, tools and data mentioned within neuroscience papers.
We propose forming a link between the two systems so that Hypothes.is can display ContentMine facts as annotations on the online document, increasing their visibility and allowing combination with manual annotations, and feedback, from the Hypothes.is user community. Certain types of Hypothes.is annotations, could also be fed back into the ContentMine facts store, providing a tight, iterative loop between people and machines.
Developing amanuens.is: the first phase
For the initial development of amanuens.is, we will focus on the growing number of open data resources that are either directly or indirectly cited in the biomedical literature. These links are often not visible to readers, and if the article is behind a paywall, they could be invisible to the vast majority of the population. We will turn data citations into intelligent identifiers.
We think that EuropePMC is an excellent underdiscovered biomedical knowledge resource and if ContentMine/Hypothes.is are successful in the first round we would:
- robustify the ContentMine pipeline and extract facts from Europe PubMedCentral’s Open Access (OA) subset to be output in a form ingestible by Hypothes.is.
- extend our capabilities to cover a longer tail of OA papers and extra open research outputs like supplementary data files.
- make use of the unique copyright exception for text and data mining in the UK to extract facts from closed publications to which we have lawful access, and expose them as open data.
-
expand our ability to mine database identifiers and find particular types of data by systematically building and adding dictionaries we can search against. Our starting point would be the list of open data sources provided by the Open Science Prize organisers.
-
output ContentMine facts as Hypothes.is annotations. As outlined above, annotators can then verify, discard as false positives or add additional information to the information uncovered by ContentMine. These would be displayed in situ on the paper and are compliant with W3C Web Annotation Working Group standards.
- most importantly, generate communities of practice. If YOU are interested in what this can do for you, then WE are interested in making it happen. That could be through adding dictionaries, starting Hypothes.is communities, or linking to key organisations.
Developing amanuens.is: the second phase
This will be significantly informed by what happens in the first. We might discover that a key technology is required, such as automating the iteration of the knowledge collection. Or maybe a set of resources - such as clinical trials - that very clearly need organizing for effective use. ContentMine are advising the OpenTrials project and delivering workshops for Cochrane UK on this topic.
By default we would initially expand to cover the whole mainstream literature and allow researchers to start mining their own custom facts. This would mean expanding the existing ability of ContentMine to limit searches to parts of a document such as the Methods section, Introduction or Acknowledgments. Want to extract your favourite DNA motifs from the results section and see what species they’re co-mentioned with? No problem. Started a new database of molecular probes and want to visualise their use in different disciplines by extracting frequency of mentions in the methods section and combining with metadata? No problem.
Two way exchange of information between scholarly communities in Hypothes.is and the ContentMine facts store would help discover more papers in a fact-centric web of knowledge and feedback looks would greatly enrich the existing pool of open data and knowledge about science on the web. We emphasize that amanuens.is is not confined to scholarly publications, but can cover government reports, NGO policies, healthcare guidelines. Indeed much of the gray literature is technically accessible - the main constraints are sociolegal.
Other partnerships and stakeholders
We are not alone in building an ecosystem for mining and both ContentMine and Hypothes.is have collaborated with the EPMC team in running joint workshops and hackathons. Peter Murray-Rust sat on the EPMC Project and Advisory Boards for 10 years and so has deep insight (and contribution) around that system. Tom Arrow at ContentMine has worked with EPMC and also with Wikimedia, forming a valuable bridge to that community along with Wikimedia volunteers Daniel Mietchen (NIH) and Magnus Manske (Wellcome Trust Sanger Centre) who have both engaged with ContentMine. Wikipedia and Wikidata are excellent resources: for the casual reader we link species, genes, chemicals directly to the Wikipedia page, but we also formally use the semantics from Wikidata, where every species, gene, place, chemical has a unique identifier. This starts to realise Tim Berners Lee’s dream of the semantic web where machines can navigate the knowledge domain. And we can also contribute! For instance, when we find a paper which is almost exclusively about a single species, we could automatically offer it to Wikidata editors for possible inclusion.
While we focus on biomedical applications, amanuens.is has almost unlimited potential. For every “Zika” we could substitute another species, another chemical, another gene. ContentMine is already organising a hackday on plant synthetic biology at The Genome Advisory Centre (TGAC), which we expect this to provide the world with new plant-oriented ontologies. Conservation initiatives such as WildLabs and Fauna & Flora International are also in contact and interested in using mining tools.
Technology and licensing
Our plan is technically viable and is a reuse and extension of open source software that already exists in alpha/beta versions with both project partners:
- Hypothes.is is already being used by scientists in imaginative ways to extract facts.
-
ContentMine is mining BioMedCentral and PLOS journals on a daily basis and has demos linking most frequently extracted facts to Wikipedia articles or other datasets like lists of endangered species.
Both systems could be incredibly more powerful and impactful if extended and combined as amanuens.is. Our proposal fits into the core missions and aims of both ContentMine and Hypothes.is and is feasible given the resources available. Linkage of the systems has been discussed for sometime and the Open Science Prize would accelerate that process and enable us to produce a more feature-rich tool than either organisation could resource independently.
All ContentMine software outputs would be made available under liberal open source licences (MIT or Apache2), Hypothes.is software is made available under the FreeBSD license with subcomponents under the MIT license. All data and annotations would be public domain under a CC0 waiver.
Supporting Information
1. An animated introduction to Hypothes.is. The Internet, peer reviewed. Available from https://www.youtube.com/watch?v=QCkm0lL-6lc .
2. Video showing ContentMine software applied to EPMC papers mentioning Zika virus (
Zika in Scientific Literature
3. An example of how amanuens.is could combine ContentMine and Hypothes.is technologies to discover information about the Zika vector Aedes aegypti and the symbiont bacterium Wolbachia (
A demo of Hypothes.is marking up a Wolbachia paper.
Project
amanuens.is
References
-
Man-Computer Symbiosis.IRE Transactions on Human Factors in Electronics1:4‑11. DOI: 10.1109/thfe2.1960.4503259
Supplementary materials
Download file (29.55 MB)