|
Research Ideas and Outcomes : Project Report
|
|
Corresponding author: Kaori L Ito (kaoriito@usc.edu)
Received: 25 Apr 2017 | Published: 03 May 2017
© 2017 Kaori L Ito, Julia Anglin, Hosung Kim, Sook-Lei Liew
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Ito K, Anglin J, Kim H, Liew S (2017) Semi-automated Robust Quantification of Lesions (SRQL) Toolbox. Research Ideas and Outcomes 3: e13395. https://doi.org/10.3897/rio.3.e13395
|
|
Abstract
Quantifying lesions in a reliable manner is fundamental for studying the effects of neuroanatomical changes related to recovery in the post-stroke brain. However, the wide variability in lesion characteristics across individuals makes manual lesion segmentation a challenging and often subjective process. This often makes it difficult to combine stroke lesion data across multiple research sites, due to subjective differences in how lesions may be defined. Thus, we developed the Semi-automated Robust Quantification of Lesions (SRQL; https://github.com/npnl/SRQL; DOI: 10.5281/zenodo.557114) Toolbox that performs several analysis steps: 1) a white matter intensity correction that removes healthy white matter voxels from the lesion mask, thereby making lesions slightly more robust to subjective errors; 2) an automated report of descriptive statistics on lesions for simplified comparison between or across groups, and 3) an option to perform analyses in both native and standard space to facilitate analyses in either space. Here, we describe the methods implemented in the toolbox.
Keywords
stroke, neuroimaging, software, neuroanatomy, robust, MRI, analysis, reliability
Introduction
In stroke neuroimaging research, reliable lesion quantification is important for performing statistical analyses relating brain changes to behavior after stroke. Specific lesion characteristics (e.g., stroke volume, type of tissues involved, and location) can provide information about inter-individual differences in post-stroke neuroanatomy. However, as conventional practice for lesion segmentation involves tedious manual tracing of lesions from structural brain images, potential inconsistencies can arise due to subjective differences in the way lesions and healthy tissue are each defined. This would be particularly problematic for large, multisite collaborative efforts that involve training of numerous individuals segmenting lesions. To improve the precision of lesion identification, we aimed to develop a reliable, user-friendly toolbox to increase the reproducibility of expert lesion segmentation and to capture lesion characteristics for statistical analysis across individuals. Here, we present the Semi-automated Robust Quantification of Lesions (SRQL; https://github.com/npnl/SRQL; DOI: 10.5281/zenodo.557114) Toolbox.
Method
The SRQL toolbox incorporates three important features to make lesion analyses more precise: (1) a semi-automated white matter intensity correction, (2) a report of descriptive statistics on lesions, such as lesion volume and lesion-to-brain volume ratio, and (3) an option to convert the lesion segmentation in native space to stereotaxic space. In this way, we are able to improve the precision and reporting of lesion segmentations.
White Matter Intensity Correction
A semi-automated white matter intensity correction was implemented to remove healthy white matter voxels that may have been inadvertently included in manual lesion segmentations (see Fig.
For each individual subject, a white matter mask is manually created in addition to the lesion segmentation. The white matter mask is a region of interest (ROI; average volume = 670±140 mm3) manually drawn on three contiguous slices from the coronal, sagittal, and horizontal planes within the healthy white matter area of the contralesional hemisphere. Both the white matter and lesion masks are then converted into binarized files. The binarized white matter mask is overlaid on the subject's T1 structural image to obtain a mean and standard deviation of the image intensity value within the region of the white matter mask. Next, the intensity of the subject's T1 structural image is applied to the binarized lesion mask to create a T1-weighted lesion mask. The value of the mean +/- one standard deviation of the white matter intensity is calculated, and applied as threshold values on the T1-weighted lesion mask. This removes voxels within the lesion mask that are within the normal intensity range of healthy white matter for that individual. Lastly, the white matter adjusted lesion mask within the subject's file is binarized as the final lesion mask for that subject.
Descriptive Lesion Statistics
The SRQL toolbox additionally outputs a single descriptive lesion database file, organized by dataset (group) and subject. For each subject, the following descriptive statistics on white matter corrected lesions are reported and stored into the lesion database file: the hemisphere of the lesion (left or right), the lesion volume (mm3), the total (skull-stripped) brain volume (mm3), and the percentage of lesion volume to total brain volume. These measurements are calculated using FSL's fslstats (
Registration to Standard Space
Finally, we implemented an option to convert the outputs (the adjusted lesion mask file and descriptive statistics) into standard space. As registration to stereotaxic space is computationally expensive, we elected to make this an optional feature for users who wish to compare lesion data across multiple subjects. To do this, the toolbox uses FSL's FLIRT toolbox (
Results
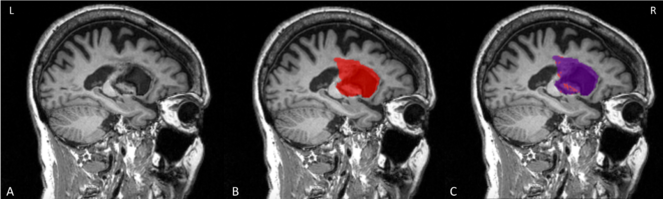
We initially tested the SRQL toolbox on a single stroke subject's anatomical scan (Fig.
Testing of SRQL toolbox on a single lesion mask. A. The stroke subject's T1 anatomical scan; B. The original lesion mask in red; C. the overlayed purple mask is the lesion segmentation after white matter intensity correction. As shown here, voxels considered to be in the healthy range have been removed by the SRQL toolbox.
We then tested the SRQL toolbox on datasets from two different sites totaling sixty subjects. Our script successfully ran on the two datasets, and our visual inspection matched the automated hemispheric categorization of each lesion. We also computed the total number of voxels removed from the original lesion volume, which was on average 4.19±12.19%. In other words, for each brain, approximately 4% of voxels fell within the healthy white matter range and were subsequently removed.
Conclusion
We developed the SRQL toolbox to improve the reliability of manual tracings and subsequently optimize the quantification of lesions across research sites. This toolbox has the ability to perform statistical analysis on lesions across multiple datasets by improving the expert’s lesion mask with white matter adjustment and reporting descriptive lesion statistics in both the original image space and stereotaxic space. By increasing the precision of lesion quantification, the SRQL toolbox lays groundwork for making greater clinical insights, such as through lesion-overlap approaches (
Future directions for the toolbox include: (1) automating the creation of each subject's white matter mask using the intensity histogram of the subject's structural image, (2) facilitating a report of group statistics on lesion data, and (3) automatically creating a lesion overlap map for visual inspection of overlapping lesion tissue across individuals.
Acknowledgements
The authors would like to thank the organizers and attendees of Brainhack LA, our research participants, and collaborators for their contributions.
Author contributions
KLI, JMA, & S-LL developed the software, performed tests, and wrote the report. HK advised the evaluation design, provided the tools for evaluation, and edited the report.
Conflicts of interest
We have no conflicts of interest to declare.
References
-
Voxel-based lesion–symptom mapping.Nature Neurosciencehttps://doi.org/10.1038/nn1050
-
A multimodal approach to understanding motor impairment and disability after stroke.Journal of Neurology261(6):1178‑1186. https://doi.org/10.1007/s00415-014-7341-8
-
A global optimisation method for robust affine registration of brain images.Medical Image Analysis5(2):143‑156. https://doi.org/10.1016/s1361-8415(01)00036-6
-
Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images.NeuroImage17(2):825‑841. https://doi.org/10.1006/nimg.2002.1132
-
FSL.NeuroImage62(2):782‑790. https://doi.org/10.1016/j.neuroimage.2011.09.015
-
Anatomy of Stroke Injury Predicts Gains From Therapy.Stroke42(2):421‑426. https://doi.org/10.1161/strokeaha.110.599340