|
Research Ideas and Outcomes : Project Report
|
|
Corresponding author: Lyubomir Penev (penev@pensoft.net)
Received: 05 Apr 2017 | Published: 05 Apr 2017
© 2017 Lyubomir Penev, Teodor Georgiev, Peter Geshev, Seyhan Demirov, Viktor Senderov, Iliyana Kuzmova, Iva Kostadinova, Slavena Peneva, Pavel Stoev
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Penev L, Georgiev T, Geshev P, Demirov S, Senderov V, Kuzmova I, Kostadinova I, Peneva S, Stoev P (2017) ARPHA-BioDiv: A toolbox for scholarly publication and dissemination of biodiversity data based on the ARPHA Publishing Platform. Research Ideas and Outcomes 3: e13088. https://doi.org/10.3897/rio.3.e13088
|
|
Abstract
The ARPHA-BioDiv Тoolbox for Scholarly Publishing and Dissemination of Biodiversity Data is a set of standards, guidelines, recommendations, tools, workflows, journals and services, based on the ARPHA Publishing Platform of Pensoft, designed to ease scholarly publishing of biodiversity and biodiversity-related data that are of primary interest to EU BON and GEO BON networks. ARPHA-BioDiv is based on the infrastructure, knowledge and exeprience gathered in the years-long research, development and publishing activities of Pensoft, upgraded with novel tools and workflows that resulted from the FP7 project EU BON.
What is ARPHA-BioDiv?
The transformation from human- to machine-readability of published content is a key feature of the dramatic changes experienced by academic publishing in the last decade. Non-machine readable PDFs, either digitally born or scanned from paper prints, require significant additional effort of post-publication markup and data extraction into a structured form, in order to address issues of interoperability and reuse of publications and data (
The next stage of development of integrated narrative and data publishing was landmarked by the Biodiversity Data Journal (BDJ) and its associated authoring tool, ARPHA Writing Tool (AWT), launched within the ViBRANT EU Framework Seven (FP7) project (
The third stage of Pensoft's effort towards open science publishing was the launch of the Research Ideas and Outcomes (RIO) journal that publishes all outputs of the research cycle, beginning with research ideas; project proposals; data and software management plans; data; methods; workflows; software; and going all the way to project reports; research and review articles, using the most transparent, open and public peer review process (
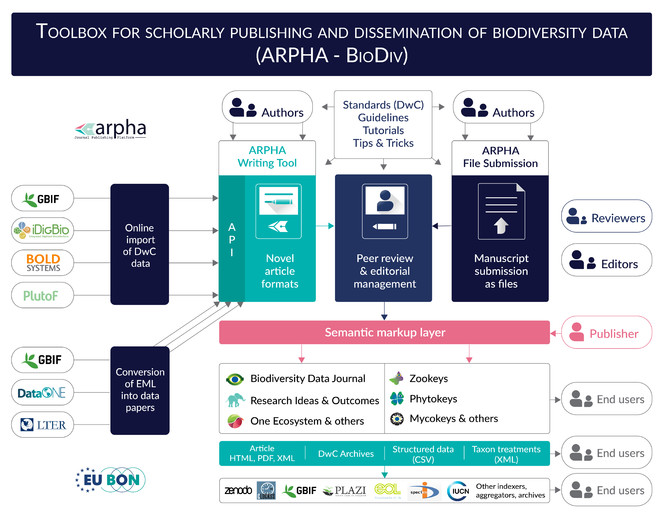
Eventually, all these years spent in development of novel approaches to publication of biodiversity data resulted in a set of standards, guidelines, workflows, tools, journals and services which we define here as ARPHA-BioDiv: A Toolbox for Scholarly Publishing and Dissemination of Biodiversity Data (Fig.
ARPHA Journal Publishing Platform
The market for online collaborative writing tools has long been dominated by Google Docs. However, as it is too generic, it has not met the specific demands of academic publishing and, in recent years, some start-ups have developed platforms and services to fulfil this increasing gap in the publishing market. Some examples include Overleaf (originally WriteLaTeX), Authorea, ShareLatex and others, most of them being based on LaTeX, but differing in the level of complexity and features for manuscript writing. For people unfamiliar with LaTeX, the learning curve is steep which explains the comparatively restricted usage, mostly centred around the LaTeX community. Currently, none of the above-mentioned tools provides all the components of an end-to-end authoring, peer review and publishing pipeline. For instance, most tools lack a peer review system and rely on integrations with well-established platforms, such as Editorial Manager, ScholarOne, or others.
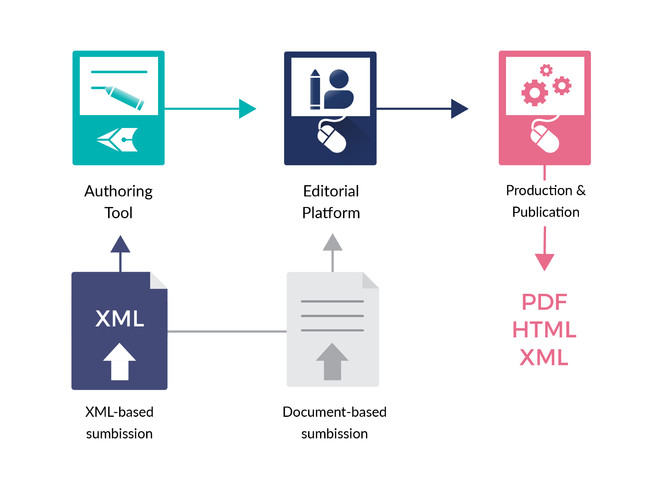
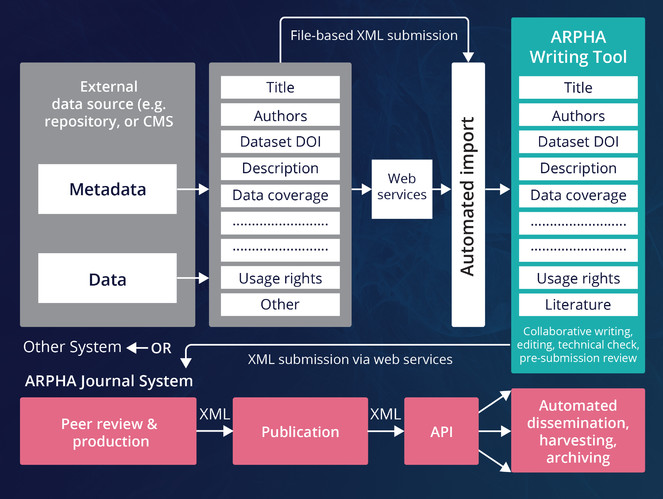
ARPHA has emerged as the first ever publishing platform to support the full life cycle of a manuscript, from authoring through submission, peer review, publication and dissemination, within a single, fully Web- and XML-based, online collaborative environment. The acronym ARPHA stands for "Authoring, Reviewing, Publishing, Hosting and Archiving" - all in one place, for the first time. The most distinct feature of ARPHA, amongst others, is that it consists of two interconnected but independently functioning journal publishing platforms. Thus, it can provide to journals and publishers either of the two or a combination of both services by enabling a smooth transition from the conventional, document-based workflows to fully XML-based publishing (Fig.
ARPHA consists of two independent journal publishing workflows: (1) ARPHA-XML, where the manuscript is written and processed via ARPHA Writing Tool and (2) ARPHA-DOC, where the manuscript is submitted and processed as document file(s).
-
ARPHA-XML: Entirely XML- and Web-based, collaborative authoring, peer review and publication workflow;
-
ARPHA-DOC: Document-based submission, peer review and publication workflow.
The two workflows use a one-stop login interface and a common peer-review and editorial manuscript tracking system. The XML-based workflow in use at Biodiversity Data Journal (BDJ) was the first of its kind back in 2013 and has since seen continuous refinement over the course of more than three years of active use by the biodiversity research community. It is also now used by the Research Ideas and Outcomes (RIO), One Ecosystem and BioDiscovery journals. The second, file-based submission workflow, is currently used by ZooKeys, PhytoKeys, MycoKeys, Journal of Hymenoptera Research, Nature Conservation, Deutsche Entomologische Zeitschrift, Zoosystematics and Evolution, NeoBiota and other journals, published by Pensoft.
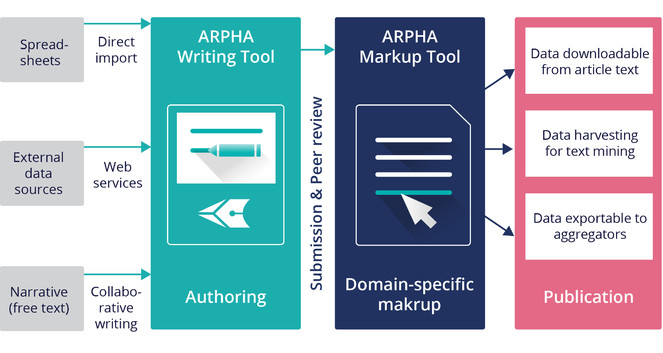
At the core of the ARPHA-XML workflow is the collaborative online manuscript authoring module called ARPHA Writing Tool (AWT). AWT’s innovative features allow for upfront markup, automisation and structuring of the free-text content during the authoring process, import/download of structured data into/from human-readable text, automated export and dissemination of small data, on-the-fly layout of composite figures and import of literature and data references from online resources. ARPHA-XML is also perhaps the first journal publishing system that allows for submission of complex manuscripts via a dedicated API.
The generic and domain-specific features of ARPHA (used for publication and dissemination of biodiversity data via the ARPHa-BioDiv toolbox) are listed in Table
Generic features of the ARPHA Journal Publishing Platform
| FEATURE | ARPHA-DOC | ARPHA-XML |
| ARPHA is a combination of software platform and a wide range of associated services. | X | X |
| ARPHA serves individual journals or multiple journal platforms. | X | X |
| Integrated with the industry leading indexing and archiving platform (see list) through web services, APIs and data exchange protocols. | X | X |
| Individual journal website design. | X | X |
| Customisable submission module. | X | X |
| Peer review and editorial management system. | X | X |
| Peer review process customisable by journal. It can be conventional (either single-blind or double-blind), community-sourced, or public. | X | X |
| Online collaborative authoring tool (ARPHA Writing Tool, abbreviated AWT, formerly Pensoft Writing Tool, abbreviated PWT), closely integrated with submission, peer review, production and dissemination tools. | X | |
| Collaborative work on a manuscript with co-authors; external contributors, such as mentors; pre-submission reviewers; linguistic and copy editors; or colleagues. The external contributors are not listed as co-authors of the manuscript. | X | |
|
Large set of pre-defined, but flexible article templates covering many types of research outcomes. |
X | |
| Online search and import of literature or data references; cross-referencing of in-text citations; import of tables; upload of images and multimedia; assembling images for display as composite figures. | X | |
| Automated technical validation step (it can be triggered by authors any time) checks the manuscript for consistency and for compliance with the JATS standard as well as the journal's requirements. | X | |
| Human-based, interactive pre-submission technical check and validation tool helps authors to proceed with their manuscripts to a form almost ready for publication. | X | |
| Pre-submission external peer review(s) performed during the authoring process. The pre-submission peer reviews are submitted together with the manuscript to prompt editorial evaluation and publication. | X | |
| For editor's convenience, peer reviews in ARPHA are automatically consolidated into a single online file that makes the editorial process straightforward, easy and comfortable. | X | |
|
In the ARPHA-XML workflow, authors can publish updated versions of their articles anytime. |
X | |
| Automated archiving of all published articles in Zenodo and CLOCKSS on the day of publication. | X | X |
Domain-specific features of the ARPHA-BioDiv toolbox used for publication and dissemination of biodiversity data
| FEATURE | ARPHA-DOC | ARPHA-XML |
|
Markup and visualisation of all taxon names used in the text. |
X | X |
| Markup and visualisation of taxon treatments following the TaxPub XML schema (an extension of the Journal Archiving Tag Suite (JATS) used by PubMed and PubMedCentral). | X | X |
| Markup and automated mapping of geo-coordinates of geographical locations. | X | X |
|
Markup and visualisation of biological collection codes against the Global Registry of Biological Repositories (GRBIO) vocabulary ( |
X | X |
| Pre-publication registration of new taxa in ZooBank, IPNI or Index Fungorum (as relevant). | X | X |
| Dynamic, real-time creation of online profile for each taxon name mentioned in an article through the Pensoft Taxon Profile tool. | X | X |
| Automated linking through the Pensoft Taxon Profile tool of each taxon name mentioned in an article to various biodiversity resources (GBIF, Encyclopedia of Life, Biodiversity Heritage Library, the National Center for Biodiversity Information (NCBI), Genbank and Barcode of Life, PubMed, PubMedCentral, Google Scholar, the International Plant Name Index (IPNI), MycoBank, Index Fungorum, ZooBank, PLANTS, Tropicos, Wikispecies, Wikipedia, Species-ID and others). | X | X |
| Workflow integration with the GBIF Integrated Publishing Toolkit (IPT) for deposition, publication and permanent linking between data and articles of primary biodiversity data (species-by-occurrence records), checklists and their associated metadata. | X | X |
| Workflow integration with the Dryad Data Repository for deposition, publication and permanent linking between data and articles of datasets other than primary biodiversity data (e.g. ecological observations, environmental data, genome data and other data types). | X | X |
|
Export of XML-based metadata and TaxPub XMLs of the papers to PubMedCentral. |
X | X |
|
Automated export of all taxon treatments (new taxa and re-descriptions, including images) to Encyclopedia of Life. Example: http://eol.org/pages/21232877/overview. |
X | X |
| Automated export of all taxon treatments (new taxa and re-descriptions) to Plazi TreatmentBank. Example: http://tb.plazi.org/GgServer/html/B07E9CD77F60DCC65C10A381F6E3BBF0 | X | X |
| Automated export of all taxon treatments (new taxa and re-descriptions), including images, keys, etc. to the Wiki repository Species-ID. Example: http://species-id.net/wiki/Spigelia_genuflexa. | X | X |
| Automated export of the occurrence data published in BDJ into Darwin Core Archive (DwC-A) format (see also |
X | |
| Automated export of the taxonomic treatments published in BDJ into Darwin Core Archive. The DwC-A is freely available for download from each article's webpage that contains taxonomic treatments data. | X | |
| Automated export and archiving of images from the published articles in Zenodo. Images from biodiversity journals are imported into the Biodiversity Literature Respository (BLR) of Zenodo. | X | X |
|
Import of Darwin Core-compliant primary biodiversity data from spreadsheet templates or via a manual Darwin Core editor and consequent publication in a structured downloadable format ( |
X | |
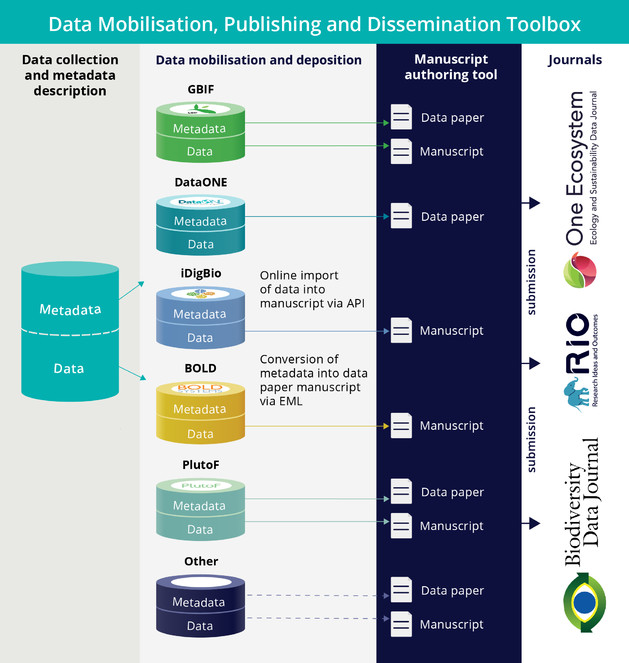
| Direct online import of Darwin Core-compliant primary biodiversity data from GBIF, Barcode of Life, iDigBio, and PlutoF into manuscripts ( |
X | |
| Multiple import of voucher specimen records associated with a particular Barcode Index Number (BIN) ( |
X | |
| Automated generation of data paper manuscripts from Ecological Metadata Language (EML) metadata files stored at GBIF Integrated Publishing Toolkit (GBIF IPT), DataONE and the Long Term Ecological Research Network (LTER) ( |
X | |
| Novel article types in ARPHA Writing Tool: Taxonomic Paper, Data Paper, Software Description, Monitoring Schema, Ecosystem Inventory, Ecosystem Service Inventory, Ecosystem Service Models, Species Conservation Profile, compliant with the IUCN Red List ( |
X* |
X |
| Nomenclatural acts modelled and developed in BDJ as different types of taxonomic treatments for plant taxonomy. | X | |
| Automated archiving of all biodiversity articles in the Biodiversity Literature Respository (BLR) of Zenodo. | X | X |
Novel Article Formats
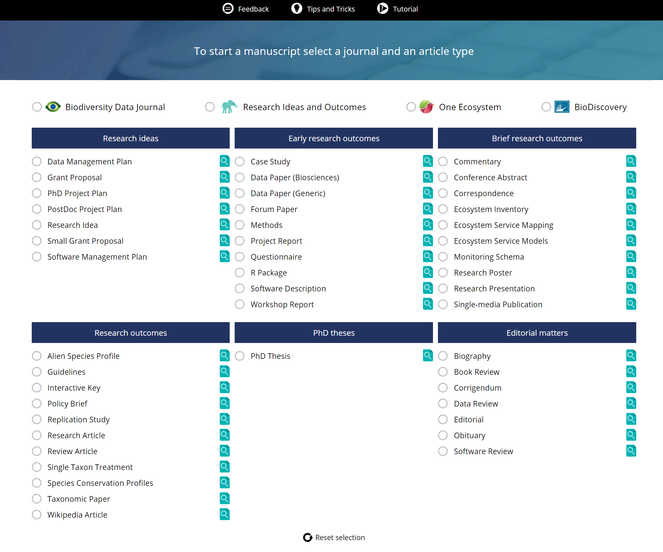
Research articles have traditionally been containers for scientiifc results for several centuries and this holds even more for research books. The Internet era brought disruptive changes to academic publishing and one of these is that the notion of the research article as the only valid output for scientific endeavours was challenged. Resulting from this, novel article formats started to proliferate in an attempt to publish extra research objects from across the research cycle, such as methods, data and software. Pensoft pioneered several novel article formats with the launch of the Biodiversity Data Journal. Currently, the ARPHA Writing Tool supports nearly fifty article formats (Fig.
Data Paper
A data paper is a scholarly journal publication whose primary purpose is to describe a dataset or a group of datasets, rather than report a research investigation. As such, it contains facts about data, rather than hypotheses and arguments in support of those hypotheses based upon data, as found in a conventional research article (for details, see
Examples from: ZooKeys, Biodiversity Data Journal, PhytoKeys, Nature Conservation.
The Article template is available for Biodiversity Data Journal, One Ecosystem, Research Ideas and Outcomes (RIO), BioDiscovery.
Software Description
A publication that describes software or an online platform. It contains a link to an openly accessible code (for details, see
Examples from: Biodiversity Data Journal.
Customisable templates are available for Biodiversity Data Journal, Research Ideas and Outcomes (RIO), One Ecosystem and BioDiscovery.
R Package
A description of an R Package including information on its purpose, installation and usage. The code should be openly available and a link to it should be present in the article.
The Article template is available for Biodiversity Data Journal, One Ecosystem, Research Ideas and Outcomes (RIO).
Monitoring Schema
A brief description of a monitoring schema including information on the monitored system component; its location; indicators used; spatial and temporal scales; purpose of the monitoring programme; and potential application of the resulting data.
The Article template is available for Research Ideas and Outcomes (RIO) and One Ecosystem.
Species Conservation Profile (SCP)
A publication of a single or multiple IUCN species assessment report(s) imported and edited in an IUCN-compliant species template.
Examples from: Biodiversity Data Journal.
The Article template is available for Biodiversity Data Journal.
Alien Species Profile (ASP)
An assessment report of alien or invasive species following an IUCN-compliant species template. After publication, the article can be exported to the Global Invasive Species Database (GISD).
The Article template is available for Biodiversity Data Journal.
Ecosystem Inventory
A brief description of a specific ecosystem type; its structures; processes and functions; abundant species; biodiversity; anthropogenic pressures; and management options. Data could result from, for example, direct observations, monitoring programmes, modelling or literature and database reviews.
The Article template is available for One Ecosystem.
Ecosystem Service Mapping
A brief description of an ecosystem service mapping study or application including information on the purpose of the map; data and methods used (biophysical, economic, social); mapped ecosystem service; mapped beneficiary (ecosystem service potential, flow, demand); spatial and temporal scale and indicators. The resulting maps should be included in the manuscript or uploaded to the ESP Visualisation Tool.
The Article template is available for One Ecosystem.
Ecosystem Service Models
A brief description of an ecosystem service mapping study or application including information on the purpose of the map; data and methods used (biophysical, economic, social); mapped ecosystem service; mapped beneficiary (ecosystem service potential, flow, demand); spatial and temporal scale and indicators. The resulting maps should be included in the manuscript or uploaded to the ESP Visualisation tool.
The Article template is available for One Ecosystem.
Semantic Tagging of the Article Content
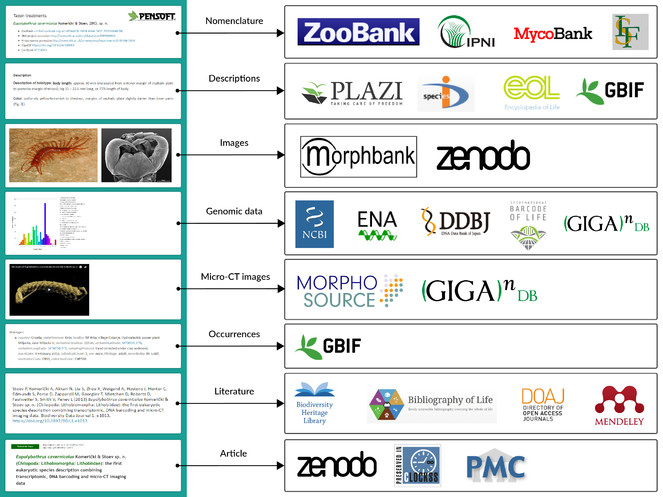
In 2010, ZooKeys published its 50th issue Taxonomy shifts up a gear: New publishing tools to accelerate biodiversity research in a new format based on pre-publication tagging of biodiversity-specific terms in the article XML and semantic enhancements to the published paper (
Examples of use of the domain-specific XML markup in the published artices.
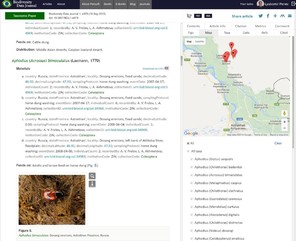
b: Pensoft Taxon Profile (PTP) is created in real time by clicking on any taxon name mentioned in an article (in this case Annoniaceae from
c: Images and pages from historic literature where a taxon name has been mentioned are available from various sources (e.g. Encyclopedia of Life and the Biodiversity Heritage Library via Pensoft Taxon Profile (PTP) (in this case Annoniaceae from
d: All taxon names usages (TNU) in an article are indexed and matched to their type of use (e.g. citations in the text, heading a taxon treatment, associated to images or present in identification keys, example from
Integrated Narrative and Data Publishing
The "integrated narrative and data publishing", or "integrated data publishing", is a relatively new approach, assuming that data or code are imported in a structured form in the manuscript text and are downloadable from the published article. In biodiversity science, this term has been coined and first demonstrated by the Biodiversity Data Journal (BDJ), developed in the course of the EU-funded project ViBRANT (
Import of Data into Manuscripts
The ARPHA Writing Tool provides online direct import from external databases using community-accepted standards (e.g. within the biodiversity community, these are Darwin Core, TaxPub JATS extension and others - see http://www.tdwg.org/standards/). Initially, data import was from CSV spreadsheets or manually via a Darwin Core HTML editor (
Another example of online import of structured text is the ReFindit tool which exists both as a stand-alone application and a plugin in ARPHA Writing Tool. ReFindit locates and imports literature and data references from CrossRef, DataCite, RefBank, Global Names Usage Bank (GNUB) and Mendeley.
Content and Data Export from Published Articles
Article content that is tagged and available in TaxPub XML can be harvested by aggregators which can select and pick sub-article elements, such as metadata, taxon treatments, occurrence records, images and others. Several of these aggregators are major players in biodiversity data preservation and management, for example, GBIF, Encycopedia of Life, Biodiversity Heritage Library, Plazi, Biodiversity Literature Repository at Zenodo, ZooBank, International Plant Names Index, MycoBank, Index Fungorum and many others. The data export in some cases is provided by a featured outbound API. The workflows and aggregators that use the semantically enriched article XMLs are listed in Table
Extraction and delivery of data and content from published articles to aggregators, nomenclators, archives, and indexers.
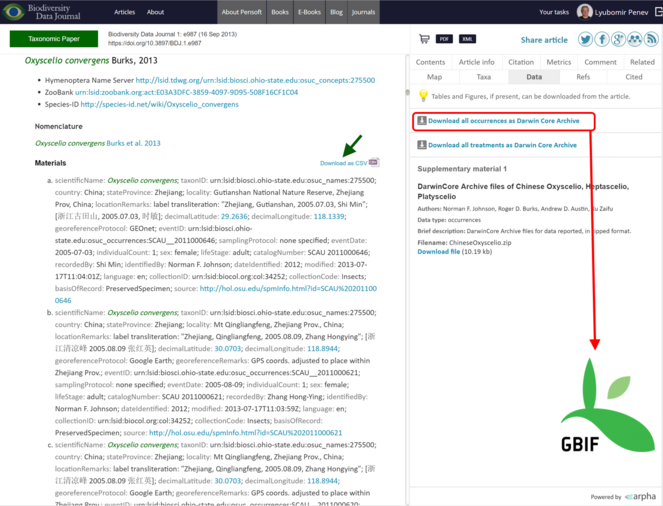
All data published in the Biodiversity Data Journal can be downloaded in tabular format (CSV) straight from the article text and re-used by anyone, provided that the original source is cited (Fig.
Export of data from articles published in Biodiversity Data Journal. Species occurrences and other structured data tables can be downloaded in CSV format (green arrow); all species occurrences are also available as Darwin Core Archives and are automatically harvested and indexed by GBIF (red box and arrow).
Data Extraction and Re-publishing Workflow
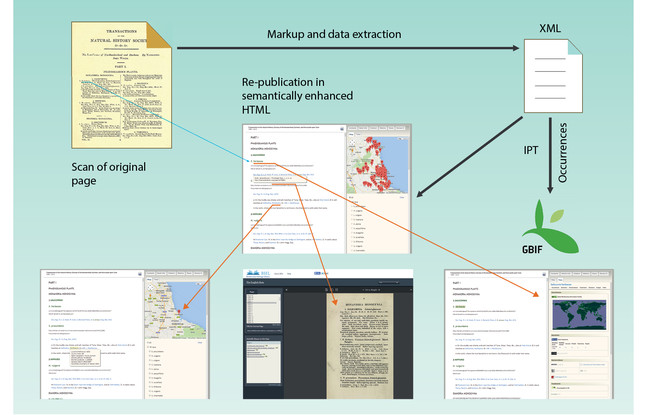
The present workflow has been created and tested with three different book titles with the support of the EU-funded projects pro-iBiospehere, SCALES and EU BON. It resulted in the launch of the Advanced Books platform of Pensoft, designed to (re-)publish historical or new books in semantically enhanced open access. The workflow is illustrated on the main homepage of Advanced Books at http://ab.pensoft.net and in Fig.
Submission of Manuscripts through an Application Programming Interface (API)
A distinct feature of the ARPHA-XML publishing workflow is the possibility to import complex manuscripts, including metadata, text figures, tables, references, citations and others, via an API available in ARPHA Writing Tool (Fig.
Submission of manuscripts to ARPHA Writing Tool through Application Programming Interface (API).
In order to submit an article via the Pensoft RESTful API, one has first to prepare an XML file according to the Pensoft XML schemas or according to the Ecological Metadata Language (EML) standard (information listed in the link above). An authentication token is obtained from the settings dialogue in the ARPHA-BioDiv which is supplied together with the XML file to the endpoint. If the document is imported successfully, it is created in the respective journal's ARPHA Writing Tool instance, where it can be further edited manually and submitted to the journal.
Creation and Publication of Data Papers from Ecological Metadata Language (EML) Metadata
Data papers, often called also “data articles”, “data notes”, or similar, were first established by the journals Ecological Archives (published by the Ecological Society of America*
The data paper should include several important elements (usually called metadata, or “description of data”), for example:
-
Title, authors and abstract;
-
Project description;
-
Methods of data collection;
-
Spatial and temporal ranges and geographical coverage;
-
Collectors and owners of the data;
-
Data usage rights and licences;
-
Software used to create or view the data.
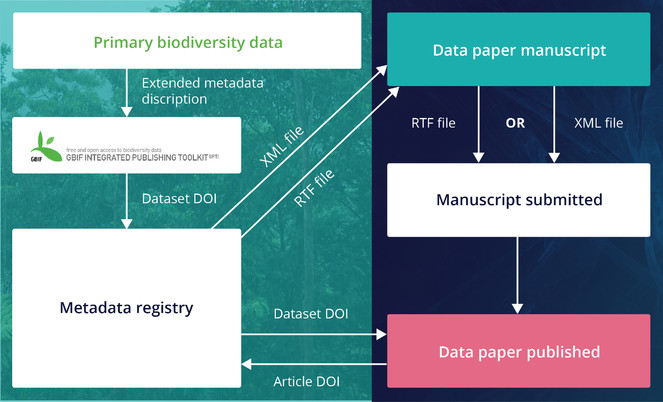
These metadata, if available and deliverable in machine-readable form (XML, JSON, etc.), can be used to produce a “data paper manuscript” that can be submitted to a journal for peer review and publication. The ARPHA approach to data paper publishing was first demonstrated in 2010 in a joint project of the Global Biodiversity Information Facility (GBIF) and Pensoft. As a result, this partnership created a workflow (Fig.
Creation of data paper manuscripts from Ecological Metadata Language (EML) metadata hosted at the GBIF IPT
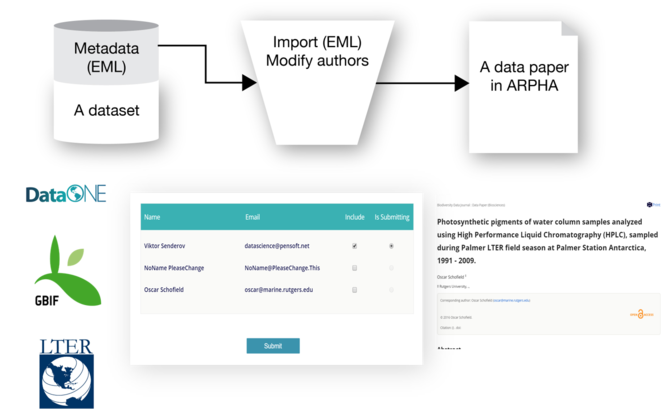
Recently, the workflow was amended by a direct import functionality of EML metadata downloadable from GBIF, LTER and DatONE networks on to a data paper manuscript in ARPHA Writing Tool (
Use Cases
The ARPHA-BioDiv toolbox has been developed in the course of several years and its tools, workflows and journals are used routinely by thousands of authors, reviewers, editors and readers worldwide. It is virtually impossible to list here the numerous use cases and approaches that have been tested and succesfully implemented over the years (see
Expert and Data Mobilisation through the Fauna Europaea Special Issue
One of the major data mobilisation initiatives realised by ARPHA and the Biodiversity Data Journal is the publication of data papers on the largest European animal database 'Fauna Europaea' within a new series "Contributions on Fauna Europaea", launched in 2014. This novel publication model was aimed at assembling in a single collection data papers on different taxonomic groups of higher rank covered by the Fauna Europaea project and accompanying papers highlighting various aspects of this project (gap-analysis, design, taxonomic assessments etc.) (
Expert and Data Mobilisation through the LifeWatchGreece Special Issue
The LifeWatchGreece special collection LifeWatchGreece: Research infrastructure (ESFRI) for biodiversity data and data observatories was published in the Biodiversity Data Journal and currently contains twenty-three papers organised in four sections:(1) Electronic infrastructure and software applications; (2) Taxonomic checklists; (3) Data papers and (4) Research articles (
EU BON Open Science Collection in RIO Journal
The journal Research Ideas and Outcomes (RIO) was designed to publish all outputs of the research cycle, from research ideas and grant proposals to data, software, research articles and research collaterals, such as workshop and project reports, guidelines, policy briefs, Wikipedia articles and others (
Guidelines, Policies and Licences for Scholarly Publishing of Biodiversity Data
Legal Framework and Policies
The legal framework and policies for publishing and re-use of biodiversity data is a subject of primary interest to the biodiversity community and policy-makers. Several EU BON teams and tasks worked on various aspects of the subject which resulted in the following set of documents:
- Open Exchange of Scientific Knowledge and European Copyright: The Case of Biodiversity Information (
Egloff et al. 2014 ) - EU BON Policy Brief on Open Data (
Egloff et al. 2015 ) - Biodiversity Data Publishing Legal Framework Report (Milestone MS841), published as a supplementary file 1 to
Egloff et al. (2016a) Egloff et al. (2016b) - Data Sharing Agreement (Milestone MS971), published as a supplementary file 2 to
Egloff et al. (2016b) - Data Policy Recommendations for Biodiversity Data (Milestone MS972) (
Egloff et al. 2016b ) - Section "Data Publishing Licenses" within the Data Publishing Strategies and Guidelines for Biodiversity Data paper (milestone MS842) (
Penev et al. 2017 )
The last two documents summarise the effort and can serve as guidelines and recommendations in the work Group on Earth Observation’s Biodiversity Observation Network (GEO BON) and beyond.
The paper of
Licences for publishing and re-use
This section from the paper of
The recommended data publishing licence used by Pensoft is the Open Data Commons Attribution License (ODC-By), which is a licence agreement intended to allow users to freely share, modify and use the published data(base), provided that the data creators are attributed (cited or acknowledged). This ensures that those who publish their data receive the academic credit that is due.
Alternatively, other licences, namely the Creative Commons CC0 (also cited as “CC-Zero” or “CC-zero”) and the Open Data Commons Public Domain Dedication and Licence (PDDL), are also STRONGLY encouraged for use in the Pensoft journals. According to the CC0 licence, "the person who associated a work with this deed has dedicated the work to the public domain by waiving all of his or her rights to the work worldwide under copyright law, including all related and neighbouring rights, to the extent allowed by law. You can copy, modify, distribute and perform the work, even for commercial purposes, all without asking permission."
Strategies and Guidelines for Scholarly Publishing
The Strategies and Guidelines for Scholarly Publishing of Biodiversity Data (
The paper also contains detailed instructions on how to prepare and peer review data intended for publication, listed under the Guidelines for Authors and Reviewers, respectively. Special attention is given to existing standards, protocols and tools to facilitate data publishing, such as the GBIF Integrated Publishing Toolkit (IPT) and the DarwinCore Archive (DwC-A).
Here, we include the table of contents of the document which will give the reader a comprehensive overview of its content (
- Data Publishing in a Nutshell
- Introduction
- What Is a Dataset
- Why Publish Data
- How to Publish Data
- How to Cite Data
- Data Publishing Policies
- Data Publishing Licences
- Open Data Repositories
- Guidelines for Authors
- Data Published within Supplementary Information Files
- Import of Darwin Core Specimen Records into Manuscripts
- Data Published in Data Papers
- Data Papers Describing Primary Biodiversity Data
- Data Papers Describing Ecological and Environmental Data
- Data Papers Describing Genomic Data
- Software Description Papers
- Guidelines for Reviewers
- Quality of the Manuscript
- Quality of the Data
- Consistency between Manuscript and Data
The Strategies and Guidelines are referred to in the Author Guidelines of Pensoft's journals and are used in their everyday publishing practices.
Tutorials, Manuals and Supporting documentation
The current article describes the rationale, overall structure and the key elements of ARPHA-BioDiv. The various elements of ARPHA-BioDiv have been featured in several papers (cited in the respective sections of the present document), guidelines, blog posts and tutorials. Below, some important supporting documentation are listed to assist the users to access this complex system.
- Overall description of ARPHA-BioDiv: ARPHA website (http://arphahub.com) and current paper.
- Guidelines for scholarly publishing of biodiversity data:
Penev et al. (2017) . - Guidelines for authors, reviewers and editors: see each journal's webpage from http://journals.pensoft.net and also
Penev et al. (2017) . - Stepwise welcome tutorial for ARPHA Writing Tool: avaiiable from the AWT menu; appears automatically to all first-time users.
- Tips and Tricks guidelines for ARPHA Writing Tool: availabe from the AWT menu.
- Promotional video for ARPHA
- Promotional video for RIO Journal
- Posts at Pensoft blog:
- Introducing the ARPHA Writing Tool
- How to import data papers from GBIF, DataONE and LTER metadata
- How to import occurrence records into manuscripts from GBIF, BOLD, iDigBio and PlutoF
- In a nutshell: The four peer review stages in RIO explained
- The 5 Most Distinct Features of ARPHA
- We ask. We listen. We innovate!
- Faster, Better, Stronger: New batch of updates now available in ARPHA Writing Tool
- Guidelines for scholarly publishing of biodiversity data from Pensoft and EU BON
Future of ARPHA-BioDiv
In the future, we want to reimagine and reinvent the academic publishing process. At the dawn of academic publishing, papers had been written especially for human consumption. The human mind alone was expected to crunch the data. Now humans rely on computers to store and manipulate the data and verify the correctness of numerical algorithms, whereas our minds focus on the big picture and the story behind the data.
With ARPHA-BioDiv, we have already taken the first few steps in creating articles that can be read both by humans and computers, as has been described so far in this article. However, more can be done. One area of innovation in academic publishing lies in creating linked content - embedding machine-readable database records in each publication that are linked to the world-wide network of linked knowledge hubs. To achieve this goal, we are currently working towards exporting content that has been semantically enriched in a knowledge graph called the Open Biodiversity Knowledge Management System or OpenBioDiv for short (
This will enable the reader of an aritcle, for example, to connect published occurrence data to portals such as GBIF and geographic repositories such as GeoNames. An illustration of the use-value of this integration will be, for example, an accelerated creation of various models, such as species distribution models, based on the article data. Thanks to the linking of the occurrence data in the article to databases, it will be possible to assemble all the elements needed for a species distribution model of the discussed taxon programmatically in an environment such as R. Moreover, the links in themselves are valuable information and can point to "hot" topics, such as "hot" taxa or "hot" figures, having many incoming links to them (
We also believe that a large portion of tradional academic publishing, even if enriched with Linked Data, will be supplemented by nano-publications (
Finally, we believe that publishers are stewards of the worlds' scientific information and there is knowledge in the totality of the published articles that is not part of any article alone. We are working on artifical intelligence algorithms both from the machine logic domain and from the machine learning domain to discover this hidden knowledge. The authors of tomorrow will have at their disposal not only a tool to format their manuscript, add citations and mark-up their data, but also tools that will discover additional information relevant to the authors' ideas and suggest similar research during the authoring phase. And, if we can dream very big, why not have artificial intelligence algorithms sophisticated enough to act as a research assistant during the authoring phase? What a marvelous thought!
Funding program
The basic infrastructure for importing specimen records was partially supported by the FP7-funded project EU BON - Building the European Biodiversity Observation Network, grant agreement ENV30845. V. Senderov's PhD is financed through the EU Marie-Sklodovska-Curie Program Grant Agreement Nr. 642241.
Author contributions
LP - vision and management; TG - technical supervision; PG, SD - software development; VS - online import of specimen records and EML metadata; IK, IK - elaboration of tutorials, promotion and PR support; SP - webdesign; PS - editorial supervision and project management.
References
-
Biodiversity data are out of local taxonomists' reach.Nature439(7075):392‑392. https://doi.org/10.1038/439392a
-
Where Do We Come From, Where Do We Go To? 20 Years Of Open Access To Biodiversity Knowledge.Zenodohttps://doi.org/10.5281/ZENODO.165979
-
LifeWatchGreece: Construction and operation of the National Research Infrastructure (ESFRI).Biodiversity Data Journal4:e10791. https://doi.org/10.3897/bdj.4.e10791
-
Linking multiple biodiversity informatics platforms with Darwin Core Archives.Biodiversity Data Journal2:e1039. https://doi.org/10.3897/bdj.2.e1039
-
Baby Killers: Documentation and Evolution of Scuttle Fly (Diptera: Phoridae) Parasitism of Ant (Hymenoptera: Formicidae) Brood.Biodiversity Data Journal5:e11277. https://doi.org/10.3897/bdj.5.e11277
-
Species Conservation Profiles compliant with the IUCN Red List of Threatened Species.Biodiversity Data Journal4:e10356. https://doi.org/10.3897/bdj.4.e10356
-
TaxPub: An extension of the NLM/NCBI Journal Publishing DTD for taxonomic descriptions.Proceedings of the Journal Article Tag Suite Conference. URL: http://www.ncbi.nlm.nih.gov/books/NBK47081/
-
The data paper: a mechanism to incentivize data publishing in biodiversity science.BMC Bioinformatics12:S2. https://doi.org/10.1186/1471-2105-12-s15-s2
-
Open_PHACTS and NanoPublications.EMBnet.journal19:17. https://doi.org/10.14806/ej.19.b.746
-
Experiences in integrated data and research object publishing using GigaDB.International Journal on Digital Librarieshttps://doi.org/10.1007/s00799-016-0174-6
-
Eu Bon Policy Brief On Open Data.Zenodohttps://doi.org/10.5281/ZENODO.188391
-
Open exchange of scientific knowledge and European copyright: The case of biodiversity information.ZooKeys414:109‑135. https://doi.org/10.3897/zookeys.414.7717
-
Copyright and the Use of Images as Biodiversity Data.bioRxivhttps://doi.org/10.1101/087015
-
Data Policy Recommendations for Biodiversity Data. EU BON Project Report.Research Ideas and Outcomes2:e8458. https://doi.org/10.3897/rio.2.e8458
-
A contribution to the study of the Lower Volga center of scarab beetle diversity in Russia: checklist of the tribe Aphodiini (Coleoptera, Scarabaeidae) of Dosang environs.Biodiversity Data Journal1:e979. https://doi.org/10.3897/bdj.1.e979
-
The Anatomy of a Nanopublication.Inf. Serv. Use30(1-2):51‑56. URL: http://dl.acm.org/citation.cfm?id=1883685.1883690
-
Creative Commons licenses and the non-commercial condition: Implications for the re-use of biodiversity information.ZooKeys150:127‑149. https://doi.org/10.3897/zookeys.150.2189
-
A nonet of novel species of Monanthotaxis (Annonaceae) from around Africa.PhytoKeys69:71‑103. https://doi.org/10.3897/phytokeys.69.9292
-
Chinese species of egg-parasitoids of the genera Oxyscelio Kieffer, Heptascelio Kieffer and Platyscelio Kieffer (Hymenoptera: Platygastridae s. l., Scelioninae).Biodiversity Data Journal1https://doi.org/10.3897/BDJ.1.e987
-
Fauna Europaea – all European animal species on the web.Biodiversity Data Journal2:e4034. https://doi.org/10.3897/bdj.2.e4034
-
Literate Programming.The Computer Journal27(2):97‑111. https://doi.org/10.1093/comjnl/27.2.97
-
Publishing the research process.Research Ideas and Outcomes1:e7547. https://doi.org/10.3897/rio.1.e7547
-
The value of data.Nature Genetics43(4):281‑283. https://doi.org/10.1038/ng0411-281
-
Editorial.The International Journal of Robotics Research28(5):587‑587. https://doi.org/10.1177/0278364909104283
-
Towards a biodiversity knowledge graph.Research Ideas and Outcomes2:e8767. https://doi.org/10.3897/rio.2.e8767
-
From Open Access to Open Science from the viewpoint of a scholarly publisher.Research Ideas and Outcomes3:e12265. https://doi.org/10.3897/rio.3.e12265
-
Implementation of TaxPub, an NLM DTD extension for domain-specific markup in taxonomy, from the experience of a biodiversity publisher.Journal Article Tag Suite Conference (JATS-Con) Proceedings.National Center for Biotechnology Information (US,Bethesda (MD)[Inen]. URL: http://www.ncbi.nlm.nih.gov/books/NBK100351/
-
Taxonomy shifts up a gear: New publishing tools to accelerate biodiversity research.ZooKeys50:1‑4. https://doi.org/10.3897/zookeys.50.543
-
Pensoft Data Publishing Policies and Guidelines for Biodiversity Data.Zenodo1:1‑34. https://doi.org/10.5281/zenodo.56660
-
Interlinking journal and wiki publications through joint citation: Working examples from ZooKeys and Plazi on Species-ID.ZooKeys90:1‑12. https://doi.org/10.3897/zookeys.90.1369
-
Strategies and guidelines for scholarly publishing of biodiversity data.Research Ideas and Outcomes3:e12431. https://doi.org/10.3897/rio.3.e12431
-
Semantic tagging of and semantic enhancements to systematics papers: ZooKeys working examples.ZooKeys50:1‑16. https://doi.org/10.3897/zookeys.50.538
-
Open Biodiversity Knowledge Management System (OBKMS).Zenodohttps://doi.org/10.5281/ZENODO.191785
-
BARCODING: BOLD: The Barcode of Life Data System (http://www.barcodinglife.org).Molecular Ecology Notes7(3):355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
-
The GBIF Integrated Publishing Toolkit: Facilitating the Efficient Publishing of Biodiversity Data on the Internet.PLoS ONE9(8):e102623. https://doi.org/10.1371/journal.pone.0102623
-
The Global Registry of Biodiversity Repositories: A Call for Community Curation.Biodiversity Data Journal4:e10293. https://doi.org/10.3897/bdj.4.e10293
-
The Open Biodiversity Knowledge Management System in Scholarly Publishing.Research Ideas and Outcomes2:e7757. https://doi.org/10.3897/rio.2.e7757
-
Online direct import of specimen records into manuscripts and automatic creation of data papers from biological databases.Research Ideas and Outcomes2:e10617. https://doi.org/10.3897/rio.2.e10617
-
Beyond dead trees: integrating the scientific process in the Biodiversity Data Journal.Biodiversity Data Journal1:e995. https://doi.org/10.3897/bdj.1.e995
-
Data publication: towards a database of everything.BMC Research Notes2(1):113. https://doi.org/10.1186/1756-0500-2-113
-
Eupolybothrus cavernicolus Komerički & Stoev sp. n. (Chilopoda: Lithobiomorpha: Lithobiidae): the first eukaryotic species description combining transcriptomic, DNA barcoding and micro-CT imaging data.Biodiversity Data Journal1:e1013. https://doi.org/10.3897/bdj.1.e1013
-
A natural language programming solution for executable papers.Procedia Computer Science4:678‑687. https://doi.org/10.1016/j.procs.2011.04.071
-
Darwin Core: An Evolving Community-Developed Biodiversity Data Standard.PLoS ONE7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
-
Flora of Northumberland and Durham.Trans. Nat. Hist. Soc. Northumberl., Durham, and Newcastle upon Tyne2:1‑149. [InPrinted by T. and J. Hodgson].
Only a part of the novel article templates (e.g. Data Papers, Software Descriptions and some others) are available in the ARPHA-DOC workflow.