|
Research Ideas and Outcomes : Project Report
|
|
Corresponding author: Cameron Craddock (brainhackorg@gmail.com)
Received: 15 Mar 2017 | Published: 20 Mar 2017
© 2018 Swati Rane, Eshin Jolly, Anne Park, Hojin Jang, Cameron Craddock
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Rane S, Jolly E, Park A, Jang H, Craddock C (2017) Developing predictive imaging biomarkers using whole-brain classifiers: Application to the ABIDE I dataset. Research Ideas and Outcomes 3: e12733. https://doi.org/10.3897/rio.3.e12733
|
|
Keywords
Machine learning, classifier, Autism, fMRI, Python
Introduction
Within clinical neuroimaging communities there is considerable optimism that functional magnetic resonance imaging (fMRI) will provide much needed objective biomarkers for diagnosing and tracking the severity of psychiatric and neurodevelopmental disorders (
Description
We implemented a Python based command-line program for training and testing disease classifiers from resting state fMRI data that was designed to be flexible enough to be run on different high performance computing platforms (e.g. distributed computing cluster). We used a modular framework based on the Scikit-learn machine-learning library (
Using the software
Running the program requires several key components: a) input directory: location of 3d NIfTI files; b) pheno_file: csv file in “long” format with subjects as rows and at least two columns containing subject identifiers and labels used for classification; c) model_dir: directory where trained models will be saved and models to be tested are loaded from; d) mask: full path to a mask file applied to each subject volume; e) model: type of algorithm to utilize. Executing the program in training mode (with the --train flag) generates a sklearn (cite) model written to disk as a serialized object, a NIfTI file containing a feature weight-map, as well as csv files containing weights at each feature, training accuracy, and model predictions.
During training, users have several options including tuning hyperparameters using a grid-search implemented via stratified five-fold cross-validation and/or imposing a sparse model solution via L1 regularization. During training the program will automatically invoke the necessary routines to: mask samples to ensure corresponding voxels are the same across subjects, reshape data into a format necessary for algorithm training, and balance label classes across training folds if hyperparameter tuning is requested. Executing the program in testing mode (with the --test flag) requires a previously trained model and saves two csv files containing model predictions and testing accuracy.
Example Use-Case: ASD Diagnostic Prediction using Regional Homogeneity:
To test our software for ASD classification, we used a preprocessed version of the ABIDE I dataset available through the Preprocessed Connectomes Project (http://preprocessed-connectomes-project.org/abide/). We specifically focused on the regional homogeneity (ReHo) fMRI derivative (
Participants and Data: The ABIDE dataset contains 539 individuals with ASD and 573 control subjects. Although most subjects were male, the ratio of males/females in both groups was identical. Gender was not considered as a feature for the classifier.
Classifier Training and Testing: First, participants were randomly divided into balanced split-half training and testing sets. During training, feature selection was performed by selecting only voxels falling within a grey matter template mask in MNI152 space. These voxels were subsequently used to train a whole brain support vector machine with L1-regularization, to enforce a sparse model solution. The hyper-parameter controlling the margin of the hyper-plane was tuned using a parameter grid-search with 5-fold cross-validation within the training set. The best performing hyper-parameter was then utilized to train a single model on the entire training set. This modeled was then applied to data from the test set in order to generate subject level predictions about diagnosis (i.e. neuro-typical or ASD). Accuracy scores were computed by comparing classifier predictions with true subject diagnoses.
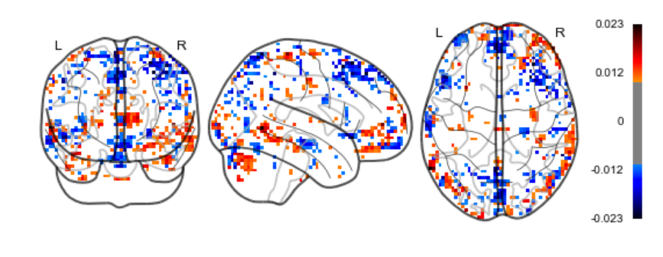
Results: Fig.
Recommendations
1. Implementation of feature selection/engineering algorithms to better develop features for predictive performance (improving speed of computation and predictive accuracy)
2. Implementation of additional al gorithms, e.g. random forest, gaussian naive bayes
Conclusions
We built a modular, python-based classification program that simplifies the model training and testing procedure for users. We then offered a proof-of-concept by using our program to predict ASD diagnoses using the ABIDE I preprocessed dataset. Using this program allowed us to build a sparse whole-brain biomarker that predicted diagnostic labels with 62% accuracy. Future improvements can include routines for feature selection and engineering, which can significantly improve computational efficiency predictive performance.
Ethics and security
All data at ABIDE I Preprocessed are fully anonymized and hence are in compliance with HIPAA.
Author contributions
CC, SR, and AP worked on conceptualization of classifier, data selection, and i/o parsing for the classifier. EJ and HJ were involved in building classifier. SR, AR, EJ, CC were involved in manuscript writing.
References
-
Deriving robust biomarkers from multi-site resting-state data: An Autism-based example.Neuroimagehttps://doi.org/10.1101/075853
-
Clinical applications of the functional connectome.NeuroImage80:527‑40. https://doi.org/10.1016/j.neuroimage.2013.04.083
-
Towards Automated Analysis of Connectomes: The Configurable Pipeline for the Analysis of Connectomes (C-PAC).Frontiers in Neuroinformatics7https://doi.org/10.3389/conf.fninf.2013.09.00042
-
The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism.Molecular psychiatry19(6):659‑67. https://doi.org/10.1038/mp.2013.78
-
Scikit-learn: Machine learning in Python.Journal of Machine Learning Research.12:2825‑2830.
-
Learning and comparing functional connectomes across subjects.NeuroImage80:405‑15. https://doi.org/10.1016/j.neuroimage.2013.04.007
-
Regional homogeneity approach to fMRI data analysis.NeuroImage22(1):394‑400. https://doi.org/10.1016/j.neuroimage.2003.12.030