|
Research Ideas and Outcomes : Project Report
|
|
Corresponding author: Julia M Huntenburg (ju.huntenburg@gmail.com)
Received: 20 Feb 2017 | Published: 23 Feb 2017
© 2017 Julia Huntenburg, Alexandre Abraham, João Loula, Franziskus Liem, Kamalaker Dadi, Gaël Varoquaux
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Huntenburg J, Abraham A, Loula J, Liem F, Dadi K, Varoquaux G (2017) Loading and plotting of cortical surface representations in Nilearn. Research Ideas and Outcomes 3: e12342. https://doi.org/10.3897/rio.3.e12342
|

|
Abstract
Processing neuroimaging data on the cortical surface traditionally requires dedicated heavy-weight software suites. Here, we present an initial support of cortical surfaces in Python within the neuroimaging data processing toolbox Nilearn. We provide loading and plotting functions for different surface data formats with minimal dependencies, along with examples of their application. Limitations of the current implementation and potential next steps are discussed.
Keywords
cortical surfaces, surface plotting, Python
Introduction
The human cerebral cortex is highly convoluted. Surface representations of neuroimaging data are essential to study cortical topography and to expose areas buried in sulcal depths. Surface-based approaches have traditionally been implemented in dedicated software suites, that can be hard to integrate with other tools. While the development of versatile Python tools for neuroimaging has recently gained momentum (e.g. http://nipy.org/), most of these tools focus on volumetric data. A notable exception is PySurfer (https://pysurfer.github.io/), a Python package for rendering neuroimaging data on the cortical surface. PySurfer provides high-level functions to visualise data processed with the Freesurfer software (
Here we present a project that departs from this landscape in two ways: it strives 1) to provide plotting for cortical surface data in Python under minimal dependencies, and 2) to integrate surface data with multivariate processing in the Nilearn toolbox (
Approach
In order to limit external dependencies to standard*
All functions are integrated in Nilearn's plotting module. The core functionality is implemented in plot_surf, which initiates the figure and axes, renders the mesh using Matplotlib's plot_trisurf function, and assigns colour for each triangle from the node-wise input data. While plot_surf provides maximal parameter flexibility, we complemented it with wrapper functions setting sensible default parameters for most common use cases.
A considerable challenge was posed by the multitude of surface file formats currently in use, and the absence of an obvious community standard. The implemented loading functions automatically determine the input type and convert it to a standard Python structure. Input can be any file that can be read by Nibabel. Internally, surface mesh geometries are represented as a list of two Numpy arrays (vertex coordinates and face indices), and data to be displayed on the mesh as a single Numpy array. It is also possible to pass these data structures directly. This design makes it easy to load common surface file formats, but also allows the user to load other formats with custom scripts.
Results and limitations
The resulting functions are demonstrated in two examples. The example data is hosted on NITRC (https://www.nitrc.org/) and data fetchers for easy download and reuse were implemented as part of this project.
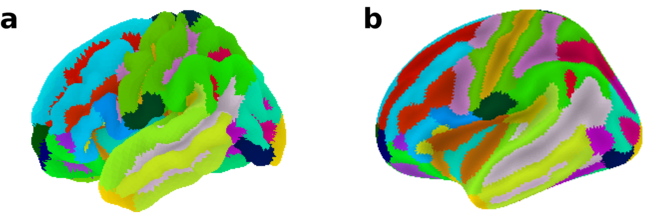
In the first example, the Destrieux atlas (
Destrieux atlas plotted on the fsaverage5 surface template using the plot_surf_roi function. a Convoluted pial surface geometry of the left hemisphere. b Inflated pial surface geometry of the left hemisphere.
The second example uses resting-state fMRI data from 1 out of 102 subjects of the enhanced NKI sample (
Seed-based functional connectivity example. a Seed region in the posterior cingulate cortex (PCC). b Pearson product-moment correlation coefficient from the seed region time series to all other nodes. c The same map as in b, thresholded and plotted with a different colour scheme. d The same map as in b, plotted without sulcal depth information for shading.
Next in the example, functional connectivity of the seed region to all other cortical nodes in the same hemisphere is calculated using Pearson's product-moment correlation coefficient. The resulting correlation map is plotted using plot_surf_stat_map (Fig.
In figures 1 and 2a-c, sulcal depth information is used for shading of the convoluted surface. While the depth data currently has to be provided by the user, it is conceivable to include utilities for calculating sulcal depth internally. If no sulcal depth information is provided, the functions default to displaying a semi-transparent mesh to expose the 3D structure without shading (Fig.
Beyond the specific limitations discussed above, some general issues remain to be solved in future work. Currently, each figure contains a single view surrounded by a lot of white space. Convenient plotting of more complex scenes, including different views and a colorbar, would be desirable. Moreover, 3D rendering remains relatively slow, a problem which is adressed in an ongoing effort to improve the underlying Matplotlib code (https://github.com/matplotlib/matplotlib/pull/6085). Finally, the present design still requires many low-level inputs from the user. To avoid this, it might be necessary to represent surfaces in a more complex object, such as a Nibabel GiftiImage. A challenge here is the lack of a standard representation of surface data in the community.
Conclusion
We implemented a set of functions to load and plot surface representations of neuroimaging data in Python and demonstrated their application in examples. The functions are easy to use, flexibly adapt to different use cases, and only require Numpy, Matplotlib and Nibabel. While multiple features remain to be added and improved, this work presents a first step towards the support of cortical surface data in Nilearn.
Acknowledgements
This work was completed during Brainhack Paris 2016 and Brainhack Anatomy Paris 2016.
Author contributions
JMH, AA and GV designed the project. JMH, AA and JL contributed to the code. KRD and GV reviewed the code. FL preprocessed the example data. JMH wrote the initial draft of the manuscript. AA, GV, JL, FL and KRD revised the manuscript.
Conflicts of interest
None declared.
References
-
Machine learning for neuroimaging with scikit-learn.Frontiers in neuroinformatics8:14. https://doi.org/10.3389/fninf.2014.00014
-
nibabel: 2.1.0.Zenodohttps://doi.org/10.5281/ZENODO.60808
-
Cortical Surface-Based Analysis.NeuroImage9(2):179‑194. https://doi.org/10.1006/nimg.1998.0395
-
The NumPy Array: A Structure for Efficient Numerical Computation.Computing in Science & Engineering13(2):22‑30. https://doi.org/10.1109/mcse.2011.37
-
Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature.NeuroImage53(1):1‑15. https://doi.org/10.1016/j.neuroimage.2010.06.010
-
Cortical Surface-Based Analysis.NeuroImage9(2):195‑207. https://doi.org/10.1006/nimg.1998.0396
-
High-resolution intersubject averaging and a coordinate system for the cortical surface.Human Brain Mapping8(4):272‑284. https://doi.org/10.1002/(sici)1097-0193(1999)8:43.0.co;2-4
-
Nipype: A Flexible, Lightweight and Extensible Neuroimaging Data Processing Framework in Python.Frontiers in Neuroinformatics5https://doi.org/10.3389/fninf.2011.00013
-
Matplotlib: A 2D Graphics Environment.Computing in Science & Engineering9(3):90‑95. https://doi.org/10.1109/mcse.2007.55
-
The NKI-Rockland Sample: A Model for Accelerating the Pace of Discovery Science in Psychiatry.Frontiers in Neuroscience6https://doi.org/10.3389/fnins.2012.00152
-
Mayavi: 3D Visualization of Scientific Data.Computing in Science & Engineering13(2):40‑51. https://doi.org/10.1109/mcse.2011.35
-
Anatomically motivated modeling of cortical laminae.NeuroImage93:210‑220. https://doi.org/10.1016/j.neuroimage.2013.03.078
where standard refers to the context of neuroimaging data analysis