|
Research Ideas and Outcomes : Grant Proposal
|
|
Corresponding author: Florian Leese (florian.leese@uni-due.de)
Received: 24 Nov 2016 | Published: 29 Nov 2016
© 2016 Florian Leese, Florian Altermatt, Agnès Bouchez, Torbjørn Ekrem, Daniel Hering, Kristian Meissner, Patricia Mergen, Jan Pawlowski, Jeremy Jay Piggott, Frédéric Rimet, Dirk Steinke, Pierre Taberlet, Alexander M. Weigand, Kessy Abarenkov, Pedro Beja, Lieven Bervoets, Snaedís Björnsdóttir, Pieter Boets, Angela Boggero, Atle Magnar Bones, Ángel Borja, Kat Bruce, Vojislava Bursić, Jens Carlsson, Fedor Čiampor, Zuzana Čiamporová-Zatovičová, Eric Coissac, Filipe Costa, Marieta Costache, Simon Creer, Zoltán Csabai, Kristy Deiner, Ángel DelValls, Stina Drakare, Sofia Duarte, Tina Eleršek, Stefano Fazi, Cene Fišer, Jean-François Flot, Vera Fonseca, Diego Fontaneto, Michael Grabowski, Wolfram Graf, Jóhannes Guðbrandsson, Micaela Hellström, Yaron Hershkovitz, Peter Hollingsworth, Bella Japoshvili, John I. Jones, Maria Kahlert, Belma Kalamujic Stroil, Panagiotis Kasapidis, Martyn G Kelly, Mary Kelly-Quinn, Emre Keskin, Urmas Kõljalg, Zrinka Ljubešić, Irena Maček, Elvira Mächler, Andrew Mahon, Marketa Marečková, Maja Mejdandzic, Georgina Mircheva, Matteo Montagna, Christian Moritz, Vallo Mulk, Andreja Naumoski, Ion Navodaru, Judit Padisák, Snæbjörn Pálsson, Kristel Panksep, Lyubomir Penev, Adam Petrusek, Martin Andreas Pfannkuchen, Craig R Primmer, Baruch Rinkevich, Ana Rotter, Astrid Schmidt-Kloiber, Pedro Segurado, Arjen Speksnijder, Pavel Stoev, Malin Strand, Sigitas Šulčius, Per Sundberg, Michael Traugott, Costas Tsigenopoulos, Xavier Turon, Alice Valentini, Berry van der Hoorn, Gábor Várbíró, Marlen Ines Vasquez Hadjilyra, Javier Viguri, Irma Vitonytė, Alfried Vogler, Trude Vrålstad, Wolfgang Wägele, Roman Wenne, Anne Winding, Guy Woodward, Bojana Zegura, Jonas Zimmermann.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Leese F, Altermatt F, Bouchez A, Ekrem T, Hering D, Meissner K, Mergen P, Pawlowski J, Piggott J, Rimet F, Steinke D, Taberlet P, Weigand A, Abarenkov K, Beja P, Bervoets L, Björnsdóttir S, Boets P, Boggero A, Bones A, Borja Á, Bruce K, Bursić V, Carlsson J, Čiampor F, Čiamporová-Zatovičová Z, Coissac E, Costa F, Costache M, Creer S, Csabai Z, Deiner K, DelValls Á, Drakare S, Duarte S, Eleršek T, Fazi S, Fišer C, Flot J, Fonseca V, Fontaneto D, Grabowski M, Graf W, Guðbrandsson J, Hershkovitz Y, Hollingsworth P, Japoshvili B, Jones J, Kahlert M, Kalamujic Stroil B, Kasapidis P, Kelly M, Kelly-Quinn M, Keskin E, Kõljalg U, Ljubešić Z, Maček I, Mächler E, Mahon A, Marečková M, Mejdandzic M, Mircheva G, Montagna M, Moritz C, Mulk V, Naumoski A, Navodaru I, Padisák J, Pálsson S, Panksep K, Penev L, Petrusek A, Pfannkuchen M, Primmer C, Rinkevich B, Rotter A, Schmidt-Kloiber A, Segurado P, Speksnijder A, Stoev P, Strand M, Šulčius S, Traugott M, Tsigenopoulos C, Turon X, Valentini A, van der Hoorn B, Várbíró G, Vasquez Hadjilyra M, Viguri J, Vitonytė I, Vogler A, Vrålstad T, Sundberg P, Hellström M, Wägele W, Wenne R, Winding A, Woodward G, Zegura B, Zimmermann J (2016) DNAqua-Net: Developing new genetic tools for bioassessment and monitoring of aquatic ecosystems in Europe. Research Ideas and Outcomes 2: e11321. https://doi.org/10.3897/rio.2.e11321
|

|
Abstract
The protection, preservation and restoration of aquatic ecosystems and their functions are of global importance. For European states it became legally binding mainly through the EU-Water Framework Directive (WFD). In order to assess the ecological status of a given water body, aquatic biodiversity data are obtained and compared to a reference water body. The quantified mismatch obtained determines the extent of potential management actions. The current approach to biodiversity assessment is based on morpho-taxonomy. This approach has many drawbacks such as being time consuming, limited in temporal and spatial resolution, and error-prone due to the varying individual taxonomic expertise of the analysts. Novel genomic tools can overcome many of the aforementioned problems and could complement or even replace traditional bioassessment. Yet, a plethora of approaches are independently developed in different institutions, thereby hampering any concerted routine application. The goal of this Action is to nucleate a group of researchers across disciplines with the task to identify gold-standard genomic tools and novel eco-genomic indices for routine application in biodiversity assessments of European fresh- and marine water bodies. Furthermore, DNAqua-Net will provide a platform for training of the next generation of European researchers preparing them for the new technologies. Jointly with water managers, politicians, and other stakeholders, the group will develop a conceptual framework for the standard application of eco-genomic tools as part of legally binding assessments.
1. Science and Technology Excellence of the Project
1.1. Challenge
1.1.1. Description of the Challenge (Main Aim)
This Action aims to advance the application of DNA-based tools and develop a roadmap to include them in the standardised ecological assessment of aquatic ecosystems in Europe and beyond, thereby optimising environmental management and improving nature conservation. Optimal environmental management is crucial, because aquatic ecosystems provide essential services ranging from clean water and filtration of pollutants to nutrient cycling, with direct impact on human society. However, land-use change, pollution, and the effects of climate change are impacting rivers, lakes, transitional and costal ecosystems and have severely degraded natural resources with negative consequences for human society. As a consequence, in 2000, the European Union implemented the Water Framework Directive (WFD) (Directive 2000/60/EC), a strict legislation to counteract degradation, initiate restoration, and manage aquatic ecosystems. The main aim of the WFD is to at least reach a good status/potential of all surface waters. The ecological status, as a main component of the overall status, is assessed through the comparison of biological quality elements (BQEs) of a given water body to undisturbed reference conditions. In four adaptive management cycles, deteriorated water bodies are identified and restored; research groups and a wide range of stakeholders are involved in the process. Although many surface waters have improved their ecological status it becomes obvious that the ambitious goals of the WFD will not be met (European Commission 2012, EEA 2012).
Although established, the bioassessment methodologies of the WFD are still intensively debated. From an application point of view, main problems associated with assessments include the variability, subjectivity and thus reliability of results, high monitoring costs, and the long time span required for sample processing; in concert these problems often allow for just one or two monitoring events per six-year management cycle (
While organisms sampled with traditional methods can be analysed in bulk, i.e. together after homogenization of tissue, there are also other approaches that make use of DNA that is present in water, sediments, or biofilms. This ‘environmental DNA’ (eDNA) can be actively captured using various molecular methods, and subsequently analysed (
Technological developments of genomic tools are progressing rapidly (e.g.
Challenge 1: Sampling protocols
Different protocols have been applied to collect the material for bioassessments. The methods comprise: (1) the WFD sampling method (multi-habitat sampling and sorting of specimens (
Challenge 2: DNA barcode references & choice of molecular biomarkers
The ultimate backbone of all molecular bioassessment protocols is the availability of a biomarker that allows the unequivocal identification of species. These markers are typically called “DNA barcodes”. For animals, the mitochondrial cytochrome c oxidase I gene (COI) is usually employed. For bacteria and protists a fragment of the ribosomal genes is targeted, while for algae and plants chloroplast coding genes are preferentially used. To date, no European DNA barcode reference database for aquatic indicators exists. This, however, is pivotal to address the WFD challenges. While many national barcoding campaigns have contributed substantial amounts of sequence data, the coverage is fragmented, incomplete and disconnected from autecological trait databases. Moreover, valid and curated DNA barcode libraries are essential to precisely identify the taxa with DNA based methods. While the use of different biomarkers for various taxonomic groups is important to obtain species level data, there is no general consensus on which amplification primers and protocols should be used to obtain optimal results for HTS. Those may be taxon and region specific and differ according to the aim of the study. For eDNA, species specific markers are needed, whereas for metabarcoding we need universal primers that capture biodiversity as a whole. These protocols will require mutual agreement on standard biomarkers and probes to ensure consistency of results.
Challenge 3: Sequencing platforms
Traditionally, the chain-termination Sanger sequencing method has been the method of choice for DNA barcoding and it is still applied in several bioassessment protocols. However, while Sanger sequencing delivers long high-quality reads and is still the standard for generating reference libraries, the drawback is that only single specimens can be processed and that per-sequence costs are high. This makes the method impracticable for high-throughput analysis, as costs are up to 3.4 times higher in comparison with the traditional morphotaxonomic assessment approach (
Challenge 4: Data analysis and storage
While every Sanger DNA barcode sequence is processed at a time by a researcher, novel HTS technologies do not allow for this level of control given that hundreds of millions of reads have to be analysed. Many different software modules and complete pipelines have been developed to perform this task automatically. However, sensitivity e.g. to the detection of sequencing errors, chimeras, or the capability of clustering sequences into distinct Operational Taxonomic Units (OTUs) varies considerably. Furthermore, monitoring based on DNA-data will generate petabytes of data that need to be stored in centralised, distributed, or semi-centralised mixed infrastructures. Thus, for routine biomonitoring a major challenge will be to identify a best-practice standard work flow to generate comparable data across monitoring intervals while ensuring long-term storage of the resulting data volume and access to High Performance Computing facilities (HPC) to perform its analysis. It is envisaged that collaborations with the European Strategy Forum on Research Infrastructure (ESFRI) and their activities such as the Partnership for Advanced Computing in Europe (PRACE, http://www.prace-ri.eu) and EUDAT (http://www.eudat.eu) will contribute to overcome this challenge.
Challenge 5: Generating novel ecogenomic indices
A central advantage of the HTS eDNA approach is that it provides information about the overall biodiversity, including small-sized organisms and cryptic species that cannot be recognised morphologically. This enormously increases the range of potential bioindicators compared to the traditional methods that are currently limited to few specific taxonomic groups such as fish, selected macroinvertebrates, and diatoms. However, little is known about the ecology of many genetically recognised species and therefore their use as bioindicators is currently limited. To overcome this problem novel ecogenomic indices based on eDNA data shall be introduced. General acceptance of such indices for bioassessment and biomonitoring will require extensive validation studies in various habitats in different European countries. Conversion and calibration with traditional methods are therefore required to allow the continuation of existing time series.
Challenge 6: Implementation of metabarcoding and eDNA methods into biomonitoring protocols and eventually into current legislation
Possibly the greatest transdisciplinary challenge in the context of this topic is to establish a framework for implementation of new methods into legislation and to involve expert committees from other disciplines involved in the WFD regulations and policy makers from the start. The current implementation plan of the WFD runs until 2027. Nevertheless, the application of genetic methods especially concerning the eDNA detection of invasive and endangered species has started already in some European countries. Therefore, there is a need for rapid Action to standardise these methods and to ensure their accuracy and efficiency so they can also be used with a broader scope in mind.
In summary, this Action will establish a framework for aquatic ecosystem assessment that uses novel genomic methods for environmental diagnostics. DNAqua-Net will bring together researchers across disciplines to identify good-practice procedures for routine applications of genomic tools and to develop novel eco-genomic indices for assessment of European water bodies. It will also provide a foundation for subsequent collaborations in the context of environmental management.
1.1.2. Relevance and Timeliness
This Action is timely for the following reasons: 1) More efficient tools are urgently needed in the context of the WFD to improve environmental management, and 2) Europe should establish itself as the global leader in environmental genomic techniques.
Coordinated strategies to counteract environmental degradation are essential for Europe and at a global scale. In order to achieve this goal a global network is needed that can assess the effects of different rehabilitation and management programs using fast, cheap, and reliable tools. With the emergence of new DNA-based techniques such as metabarcoding and eDNA screens, such tools are increasingly available. More importantly, they can be easily standardised and compared with ongoing traditional assessment routines. Thus, complementing ongoing environmental assessment programs with such tools to counteract biodiversity loss is of utmost relevance. This has already been realized by the US Environmental Protection Agency (EPA), which has implemented a roadmap for the inclusion of DNA-based methods in water quality assessments since 2011 (http://www.epa.gov/eerd/research/dnabarcoding.html). However, focus was primarily on traditional Sanger sequencing based assessment methodology (
1.2. Objectives
1.2.1. Research Coordination Objectives
DNAqua-Net has a pan-European focus on fresh- and marine waters and connects to International Partner Countries (IPCs) and Near Neighbour Countries (NNCs) to achieve the following Research Coordination Objectives:
1) Nucleating existing knowledge. For some of the research challenges there are already possible solutions being tested in individual labs. The first objective of DNAqua-Net is to provide a platform for coordinated discussions about good-practice examples.
2) Method comparison: Through the network, a standardised comparison between different methodological approaches, as well as between classical and DNA-based methods will be performed to evaluate and quantify the innovation potential for bioassessment.
3) DNA barcoding reference data bases at the European level: Current data bases are lacking many indicator taxa. Thus, there is a need to identify gaps through a Europe-wide data comparison.
4) Bridging gaps between disciplines. DNAqua-Net will connect researchers involved in the development and application of the traditional aquatic assessment methods with researchers applying DNA-based tools to discuss gaps and solutions for a novel bioassessment and monitoring system ("WFD 2.0").
5) Dissemination of research. Information on the potential and pitfalls of the different techniques will be disseminated as Standard Operational Procedures (SOPs) or good-practice strategies to research communities, stakeholders and the public. The task will be addressed by individual Working Groups (WGs) and will find entry into open access review/opinion papers and guidelines for stakeholders that are published during and beyond the timeframe of the Action.
6) Data-storage and HPC concept. The problem of Big Data analysis and storage for genomic bioassessments is apparent. DNAqua-Net will develop guidelines for HPC data analysis as well as long-term handling and accessibility of these data.
7) Input for future market applications. The project will contribute to economic development by providing opportunities to create new start-ups specialised in ecogenomic analyses and by developing new capacities in existing environmental agencies and consulting companies.
1.2.1. Capacity-building Objectives
DNAqua-Net seeks to drive both scientific progress and technological innovation in this highly relevant field of research through the following Capacity-building Objectives:
1) Fostering knowledge exchange in the emerging field of ecogenomic bioassessment.
2) Connecting researchers and stakeholders involved in current bioassessment procedures with researchers developing novel genomic techniques.
3) Establishing a platform including a web-based system to inform stakeholders about the new techniques and gold-standard approaches for their application.
4) Facilitate access to workshops, Short-Term Scientific Missions (STSMs), and conferences especially for Early Career Investigators (ECIs), under-represented gender, and participants from countries with less developed structures for genomic analyses through the Action’s budget.
5) Inclusion across EU-countries: DNAqua-Net actively involves institutions from new EU-13 and old member states already from the stage of application (proposers from 8 old and 4 new EU-13 member states).
1.3 Progress beyond the state-of-the-art and Innovation Potential
1.3.1. Description of the state-of-the-art
Different biomarkers have been accepted as DNA barcodes for animals (
The applications of DNA barcoding in ecology range from species detection to biodiversity assessments and the analyses of species interactions. A particular effort has been made to use eDNA for genetic monitoring of invasive and endangered species (
From the perspective of bioassessment and biomonitoring, the two main directions of eDNA research are the detection of targeted species and the assessment of diversity of particular bioindicator taxa. While the first line of research is mainly based on Polymerase Chain Reactions (PCR) or real-time and digital droplet PCR analyses (
1.3.2. Progress beyond the state-of-the-art
DNA-based methods have the potential to fundamentally improve biomonitoring and thus nature conservation. However, prior to the concerted implementation of the technology in ecological assessment (Challenge 6) there needs to be a central agreement on workflows, ranging from sampling and lab protocols to the calculation of biotic indices with highly standardised bioinformatics pipelines (Challenges 1-5). To this end, this Action network will bring together leading scientists representing both the innovative and more traditional monitoring approaches for a dialogue on how to standardise and further improve the innovative technology and to discuss a conceptual framework for its implementation in biomonitoring. It will act as a “think-tank” catalysing dialogue on the scientific basis of the rapidly advancing technology through meetings, a communication platform, opinion and review papers, and STSMs. STSMs will allow lab visits between EU-countries, with a particular emphasis on connecting new and old EU member states and on providing opportunities to test methods through available funds of national projects.
1.3.3. Innovation in tackling the Challenge
In the light of aforementioned challenges, such as the gaps between the emerging potential of new HTS technologies and lack of their wide application, DNAqua-Net is innovative and timely, as it seeks to combine the largely disconnected knowledge among institutions to identify good-practice protocols for environmental monitoring. Benefits of this project go far beyond academic results: A main scientific innovation of DNAqua-Net will be the modification of existing biotic indices to fit the specificity of eDNA data and the development of new eco-genomic indices for ecosystem assessment. This process will require a set of new molecular and analytic tools to acquire and analyse the genetic data. The conceptual development is part of this Action. Furthermore, through intensified collaboration and a central access point and portal, DNAqua-Net will specifically advance DNA-barcode reference library compilation by identifying and closing existing gaps across countries. Thereby, DNAqua-Net will contribute to ongoing large-scale DNA barcoding initiatives that compile databases serving as backbone for eDNA/metabarcoding analyses.
Another key innovation of DNAqua-Net is the connection of molecular ecologists with researchers involved in traditional bioassessment as well as with stakeholders on a European scale and beyond. Through this, the Action will identify a process how to “translate” traditional and DNA-based bioassessment results into each other and how to implement the new methods into practical biomonitoring encompassing the best approaches of both. It will further initiate pilot applications. The Action network will provide training, guidance, and intercalibrated protocols on how to apply novel and fast ecogenomic indices (or markers) in bioassessments to reliably identify and quantify environmental degradation and thus help natural resource management programs. Therefore, DNAqua-Net has direct and significant implications for societal policy because management decisions as well as conservation and restoration plans can be based upon and monitored using highly standardised and much more comprehensive data on aquatic biota and associated functions.
1.4. Added Value of the networking
1.4.1. In relation to the Challenge
In the EU, water quality assessment is a mandatory pan-European challenge regulated by different Directives (e.g. Water Framework Directive 2000/60/EC, Habitats Directive 92/43/EEC, Marine Strategy Framework Directive 2008/56/EC). To fulfil the demands of the Directives, networks of ecologists, water managers, and policy makers have developed strict implementation plans. The added value of this Action is to initiate a pan-European network of researchers using DNA-based methods applied to water quality assessment and link this new network to existing ones. The specific value of DNAqua-Net is thus that it brings together molecular biologists, taxonomists, ecologists with water managers, policy makers, economy, and stakeholders. Direct communication is absolutely essential to integrate available knowledge on field/lab protocols, bioinformatics analysis, biomarkers, indices, and sequencing platforms used (Challenges 2-5) and pull together information on DNA barcodes of relevant indicator taxa (Challenge 1). Furthermore, this Action will stimulate long-term research and technological development. Within the context of the WFD, the network will lead to faster adoption of standardised protocols in the implementation of River Basin Management Plans and lead to considerable gains in management efficiency (Challenge 6). Further, the results of this Action will be of utmost relevance for other Directives involved in the management of biodiversity (e.g. Bird Directive 2009/47/EC and 79/409/EEC, the Groundwater Directive 2006/118/EC and in the context of the development of a new Soil Directive for the EU). Ultimately, DNA-based techniques developed through the Action network have the clear potential to provide more cost efficient and rapid tools to assess the effect of costly restoration and conservation programs.
1.4.2. In relation to existing efforts at European and/or international Level
This Action will move forward the existing pan-European efforts to develop an EU-wide strategy for the improvement of biomonitoring. At present, national and international Barcoding initiatives are compiling lists of their respective national inventories of organisms (see above and references). There are, however, only limited efforts to specifically focus on the target taxa needed to address the WFD challenges, such as diatoms or benthic invertebrates. Here, networking through this Action will bridge the gap by comparing the national operational taxa lists for WFD monitoring to the available barcode databases. Specifically, the species not represented in the databases shall be obtained and studied. This research objective connects to national Barcode projects, the international BOLD reference library, as well as several prominent EU-wide Earth observation programes (e.g. Natura 2000, LTER-Europe, EU-BON) and thus creates synergies between these and DNAqua-Net. None of the aforementioned programs uses DNA-based methods to evaluate the quality of Europe’s waters. Through networking inside DNAqua-Net as well as linking to other networks, this Action will address novel and highly relevant topics going far beyond current European or international programs.
2. Impact
2.1. Expected Impact
2.1.1. Short-term and long-term scientific, technological, and/or socioeconomic Impacts
The overarching objective of DNAqua-Net is to establish a cross-discipline, international network of scientists, managers, governmental institutions, manufacturers, and emerging service providers to identify major challenges in DNA-based bioassessment and provide standardised best-practice solutions to those.
Short-term impacts
Short-term scientific impacts are i) the harmonisation of existing knowledge and an advancement of DNA-based monitoring techniques, connecting research institutions (particularly in new and old EU countries), ii) the advancement of DNA barcoding projects and campaigns in order to complete existing reference databases, iii) provision in training for HTS methods, and iv) the initiation of national pilot projects parallel to traditionally performed assessments to compare performance and to guide further scientific and technological development.
Technological impacts lie in the dissemination of new lab protocols and bioinformatics tools to analyse HTS metabarcoding data across working groups / member states.
Short-term socioeconomic impacts will likely be the encouragement of manufacturers to provide adequate tools and the emergence of start-ups providing ecogenomic services for ecological assessment (biomonitoring). DNAqua-Net will also involve amateur taxonomists in the process of collecting and determining samples for the reference barcode library. In addition, the general public will be involved in the dissemination of the results of the Action network to raise awareness of water quality issues. This will be done through citizen science events such as Action days at which members of DNAqua-Net show how the new assessment methods work in real world scenarios.
Long-term impacts
Given the recent technological advancement of DNA-based methods it is expected that these techniques will gradually replace traditional field and lab procedures in bioassessment over the coming ten to fifteen years. In the EU, DNA-based aquatic ecosystem monitoring for the WFD is likely to advance most rapidly, due to the existing well developed infrastructure of monitoring networks (e.g. more than 100,000 sampling stations for rivers alone but also for lakes, transitional and coastal ecosystems). Replacing traditional labour intensive sampling, sorting, and identification with more reliable and eventually also cheaper methods will (i) greatly enhance the quality of data used for decision making in the protection of aquatic resources and (ii) save considerable public resources. By providing a forum for scientific discourse, DNAqua-Net also sets the stage for long-term international transdisciplinary collaborations that will result in joint research proposals that could not have been initiated without this Action. Therefore, the outcomes of this Action will endure beyond the proposal’s four-year period. Further long-term impacts will be the implementation of a forum connecting researchers working on the new genetic technologies with stakeholders and researchers involved in traditional bioassessment approaches. This will ensure the implementation strategy of genetic techniques becoming part of ongoing monitoring cycles of the WFD especially for the time after the current legislation frame (2000-2027), i.e. an implementation plan for a “WFD 2.0”. These discussions will be in line with the EU strategy referred to in the “Blueprint to Safeguard Europe’s Water Resources” (EC 2012) and it’s long-term vision (until 2050). A further advantage of the formed pan-European DNAqua-Net scientific network is that research can be performed more cost-efficiently due to the synergies through collaborations.
The enduring impact of this Action will be the establishment of European leadership in the field of DNA-based assessment of aquatic ecosystems. Monitoring systems for rivers, lakes, transitional water and coastal ecosystems exist in most parts of the world and many countries have adopted methods comparable to the WFD. Therefore, there is substantial potential for harmonising DNA-based methods and consequently improving aquatic monitoring worldwide. If European companies and research institutions can establish themselves as leaders in this field there will be immense market opportunities to create jobs in different sectors ranging from SMEs to larger industry and academia. A vital precursor for European enterprises to gain a competitive edge in the eDNA and DNA-based bioassessment market is the set of standardised protocols envisaged in this project. The standardisation of DNA-based bioassessment will help to unify the market and increase overall market volume for manufacturers of HTS equipment and preparatory devices. The resulting specialist equipment will enable staff with minimal training to perform bioassessment tasks formerly restricted to highly qualified scientists. This will create business opportunities for start-ups and expanding opportunities for existing bioassessment SMEs. Overall, the resulting novel technologies and products have a strong potential to create new jobs for lesser qualified people while concurrently enabling highly qualified personnel to switch from routine to expert bioassessment tasks. Standardised eDNA tools will also be of utmost importance for the use in other contexts, such as i) the compilation of IUCN red lists, ii) invasive species programs, iii) food chain trackers, iv) agriculture and v) forestry (e.g. FSCs). Thus, the outcome is expected to contribute directly to nature conservation at larger scales.
2.2. Measures to maximise Impact
2.2.1. Plan for involving the most relevant Stakeholders
The results of the project will be of special relevance to stakeholders related to the implementation of the WFD, Habitats Directive and Marine Strategy Framework Directive (MSFD). While not all developed methods will be immediately applied on a pan-European scale, it is our aim to introduce developed protocols in the CEN standardisation process as new working item proposals (NWIP) within the duration of the project. Thus, pilot applications of this COST Action will become standardised CEN guidance which will ensure their application in the 33 CEN member and 17 associated countries in the near future. Further, many CEN standards may also be adopted as ISO standards, especially if comparable ISO guidance is not yet in place, as is the case here.
To reach these goals, the approach of DNAqua-Net towards stakeholder involvement is twofold: First, a cooperation in detail with a number of stakeholders representing the different levels of WFD and MSFD implementation (river basin management, marine coastal bioassessment, national and EU-wide implementation) will be initiated as part of pilot applications. Here, specific contacts have already been made with the International Water Assocation (IWA), the European Water Association (EWA) and their national branches as well as with the CEN SABE team dealing with strategic issues (i.e. Environmental Monitoring Strategy Team, ENV). Secondly, DNAqua-Net will involve and expand the existing range of stakeholders from EU countries and beyond through inclusive involvement and participation, which will sharpen the project’s outcomes and will lay the foundation for broad application in the future.
For both, the detailed and the more general level of involvement of these partners are included:
- River basin authorities, being responsible for the practical monitoring activities (including lakes, transitional and coastal waters), as well as companies and consulting agencies performing the monitoring. For the detailed level representatives of several catchments will be included, covering different regions and different levels of taxonomic knowledge.
- National authorities responsible for the nation-wide coordination of monitoring programs and the data delivery to the EU. These authorities will finally be in charge to adopt new field and lab methods and to ensure the comparability of the results to previous monitoring programs. It is therefore crucial to collect their opinions and recommendations.
- CEN SABE to improve exchange between science, industry, and policy communities. Involving CEN SABE ENV will encourage a more rapid uptake of the new technologies and methods in standards and ensure a better understanding of the demands of industry and end-users who have to fulfill environmental requirements.
- The relevant bodies of the Common Implementation Strategy (CIS) of the WFD being responsible for the Europe-wide coordination of WFD implementation http://ec.europa.eu/environment/water/water-framework/objectives/implementation_en.htm. Within the currently adopted CIS structure, the Working Group ECOSTAT is the relevant group, considered as the main Europe-wide stakeholder. Given the continuous evolvement of the CIS process, other groups might need to be included in future. ECOSTAT will primarily solve questions related to the intercalibration of assessment methods to ensure a harmonised application of monitoring throughout Europe. To involve the relevant partners internationally, EWA as well as IWA and the European Innovation Partnership on Water (EIP Water) will also be central contact points of this Action.
- Certified Laboratories currently involved in monitoring water quality, providing species lists according to the WFD.
- Citizens and users of water. DNAqua-Net will involve the public, including water companies, managers of water bodies, fisheries, and amateur taxonomists through Action days, during which sampling and analysis are demonstrated in situ to raise awareness of the necessity of ecological assessments and the power of new approaches.
From the beginning, DNAqua-Net actively involves many researchers of the inclusiveness target countries to achieve the Action’s aim, the improvement of environmental management across Europe and beyond. Through funds specifically assigned to Early Career Researchers and underrepresented gender, DNAqua-Net will promote their participation in this Action.
2.2.2. Dissemination and/or Exploitation Plan
To foster collaborations and disseminate knowledge, DNAqua-Net will primarily make use of the Action programs. These will include about 16-24 STSMs, 4 Training Schools in HTS and bioinformatics analysis, 4 workshops, 4 annual meetings and two international conferences with invited speakers from COST member states, NNC, and IPCs. DNAqua-Net will make all results of WG meetings, workshops, and conferences publicly available. Aside from traditional channels (publications, web entries), the Action will actively invite stakeholders to meetings and use novel media (Blogs, Webinars, e-Learning outreach, Twitter, Facebook, Youtube) as well as the public outreach offices of the different institutions to maximize the dissemination of knowledge.
Two large high-profile conferences in the first and last year of the Action will be widely advertised. They will attract a diverse audience, including ecologists, representatives of environmental offices, and governmental agencies involved in the legislation as well as the CEOs of private companies involved in environmental monitoring. A joint organisation together with EWA or IWA is considered to maximise impact and dissemination.
Reports: The five WGs will also produce reports on specific topics (e.g. good-practice guides for different tasks) as soon as agreement has been reached. These will be disseminated online and through social media, as well as through public relation offices of institutions.
‘Cookbook’ and CEN new working item proposals (NWIP): The production of a so called ‘cookbook’ with details of suggested protocols for the accurate ecogenomic assessment of the ecological state in different aquatic ecosystems will be organized by the Core Group. It is expected that scientists from NNC and IPC institutions will collaborate on the production of the cookbook. Developed protocols and methods fit for standardisation will be introduced to the CEN standardisation process as new working item proposals. The cookbook as well as the CEN NWIP are expected to be available before the end of the Action (year 4).
Open Data and Open Science policy: One of the objectives is to provide and promote a barrier-free clear picture of the current state of knowledge and proposed research priorities. In order to make these conclusions widely available among the scientific community, open-access peer-reviewed publications in the form of opinions and reviews as well as method papers will be produced; these publications will be advertised on the Action website, and are expected to be available before the end of the Action (year 4). Sequences generated via Barcoding projects also as part of STSMs will be submitted to GenBank and BOLD, species lists will be shared with GBIF, LTER, and the GEO BON portal.
2.3. Potential for Innovation versus Risk Level
2.3.1. Potential for scientific, technological and/or socioeconomic Innovation Breakthroughs
Innovation
The scientific and commercial innovation potential of HTS methods is enormous. A fast growing number of publications in molecular ecological research aptly document the potential of the technology for rapid and reliable assessment of ecosystems. Furthermore, they allow addressing additional questions such as responses of communities, species, or genotypes to multiple stressors, while allowing inference of foodweb structure and genetic diversity from species to biomes. Thus, environmental genomic tools have a clear application range beyond ecosystem assessment or species monitoring in aquatic ecosystems. The tools can be used at global or local levels (IUCN red lists, FSC forestry validation, validation of the content of traditional Chinese or other medicines, species validation at customs and more). Therefore, the technological breakthrough of novel environmental genomic tools shows immense promise and opportunity.
The new HTS methods and monitoring approaches offer the potential to enhance European technological leadership and to establish or improve the business of globally acting companies in the field of environmental monitoring. A key role in promoting economic impact of DNAqua-Net will be the standardisation of project findings as CEN guidance. This will both grow and establish a market for manufacturers of HTS equipment as well as SME service providers of HTS-based bioassessments. DNAqua-Net deliverables will also help environmental agencies to improve their surveillance capabilities for detrimental human activities thus protecting both environmental and human well-being. The expected continuation of rapid development in the sector of HTS techniques will lead to more cost effective and accurate bioassessment, benefitting both the corporate and public sectors. Expected improvements in accuracy and prediction will result in more effective use of public money in aquatic ecosystem remediation actions.
Risks
Compared to the documented potential of innovation, the risks of novel DNA-based approaches are restricted to a few primarily technological aspects.
i) Standardisation of workflows might be impeded by continuous development of DNA sequencing technologies creating new sequencing platforms every 5-10 years. Solution: it is necessary to formulate SOPs in such a way that they deliver comparable data even if platforms change. In addition, connectivity of novel economic and traditional biodiversity assessment methods needs to be possible.
ii) Implementation strategy: The majority of agencies and institutes involved in biomonitoring using traditional methods lack the technical facilities and expertise for molecular analyses. Solution: Through open workshops and an active dissemination strategy, this Action will involve the relevant institutions and offer training in how to obtain and interpret the data.
iii) Conformity of novel data with EU legislation: While DNA-based tools have already a tremendous resolution in identifying endangered species and invasive species and also provide exceptionally detailed lists on species diversity in ecosystems, they are limited in their robustness to provide biomass data. Currently, the WFD demands this information for some national indices on BQEs which may delay the acceptance for DNA-based indices within the WFD context. The proposers are aware of research efforts to address this issue and, based on emerging studies, confident that technological innovation will provide solutions to this issue within the next few years.
3. Implementation
3.1. Description of the Work Plan
3.1.1. Description of the Working Groups
DNAqua-Net consists of the five following Working Groups (WGs) that implemented in accordance with the identified challenges for this Action:
WG 1: DNA Barcode Reference Databases
The aim of this working group is to identify gaps for aquatic species in ongoing national DNA barcoding projects. Specifically, the WG members aim at filling these gaps in cooperation with the partners of the pan-European network. Funding for these activities is available as part of national and international Barcode of Life projects (e.g. GBOL, NorBOL, SwissBOL etc.). DNAqua-Net will provide an efficient web-based platform to inform on missing species needed for the taxon lists used in different biomonitoring activities (WFD, MSFD). Also, this WG will transfer knowledge on how to setup national DNA-barcoding campaigns specifically for many Inclusiveness Target Countries (ITC) that have no such activities. A milestone in the first year is the setup of a website that lists important taxa for traditional biomonitoring that are not included in Barcode databases yet. Based on this list, a targeted outreach will be launched to collect and sequence in order to improve reference libraries. The WG will organise two workshops on DNA barcoding and data management. Furthermore, several STSMs will be advocated in order to fill the gaps in the databases by connecting traditional morphotaxonomists to DNA barcoders and staff of BOLD, GBIF, LTER-Europe, GEO-BON, and other projects and institutions.
WG 2: Biotic Indices and Metrics
The aim of this WG is to assess the ecological value of potential new bioindicators identified by environmental DNA barcoding and to discuss the revision of currently used biotic indices to make them better adapted to the specificity of HTS data. In the context of ongoing monitoring, the group will compare available data of genetic assessments with traditional assessments. The WG will also evaluate the possible introduction of novel ecogenomic indices and the conditions of their validation. The results shall be made available through WG reports but also specifically as Opinion Papers in peer-reviewed journals. Definitions for a central database and architecture will be formed. A close cooperation with WGs 3-5 will be sought.
WG 3: Field and Lab Protocols
The aim of this WG group is to discuss and evaluate current protocols for sampling, processing, and laboratory approaches with the goal of identifying gold-standard field and lab protocols to assess BQEs with eDNA/metabarcoding. The WG will organise workshops for the assessment of whole aquatic biodiversity (bulk / eDNA metabarcoding) as well as techniques for detecting rare/endangered or invasive species (real-time PCR, digital droplet PCR). Furthermore, training workshops will be used to inform on guidelines for performing these techniques in routine application. The group will deliver reports of the proposed solutions published on the Action website and through an associated entry in the ‘Cookbook’. Furthermore, several STSMs will be coordinated with WGs 2-4.
WG 4: Data Analysis and Storage
The main aim of this WG is to identify best-practice approaches for data analysis. The group will work in close cooperation with WG 3 to understand the requirements for the input materials. In addition, it will work in close cooperation with WG 2 to implement the conceptual framework of quantifying biodiversity with the ecogenomic index in the new pipeline. A central aim of the WG will be to connect with other HPC / Big Data EU-projects such as PRACE and EUDAT, and discuss if solutions found in these projects are applicable to the HTS data problem. The WG will organise at least two training schools for HTS data analysis and data management. Furthermore, several STSMs will be coordinated between WGs 2-4. There will be a report proposing the structure of a bioinformatics pipeline using current resources, platforms, and databases but also a definition of functionalities will be developed.
WG 5: Implementation Strategies and Legal Issues
This highly transdisciplinary WG consists of several researchers involved in WGs 1-4, researchers involved in the traditional WFD assessment scheme and in particular stakeholders and policy makers. The aim is to develop a conceptual framework based specifically on the results of WG 2-4 for new methods for real-world and large-scale assessments. Therefore, a first milestone during the first WG meeting is to learn what type of data and protocols water managers need in order to provide WGs 2-4 with such information. Special emphasis shall be paid onto legislation aspects that are critical to discuss and plan on a long-term basis. The WG will also actively advocate pioneering projects as part of the fourth management cycle to run side-by-side with traditional methods. 3-4 training schools will be organized in different countries across Europe by this WG.
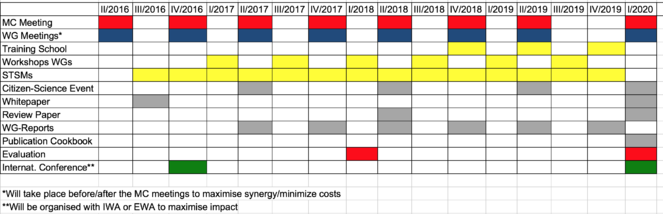
The proposed timeline for the Action is shown in Fig.
Year 1 Milestones
- 1st Management Committee (MC) meeting and 2 WG meetings
- International Action conference (inauguration of DNAqua-Net)
- 2nd MC meeting / Core Group (CG) meeting
- Setup of the Action Website (www.DNAqua.Net)
- 4-6 STSMs
- Action workshops (HTS techniques, DNA barcoding, Bioinformatics)
- White Paper published by the CG
Year 2 Milestones
- 2 MC/CG and WG meetings (taking place shortly before or after MC meeting)
- 4-6 STSMs
- Joint Student Supervision (combined with STSMs)
- Action workshops (HTS techniques, DNA barcoding, Bioinformatics)
- 4 WG reports
- Mid-term evaluation report
- Review paper
Year 3 Milestones
- 2 MC/CG and WG meetings (shortly before or after MC meeting)
- at least 4-6 STSMs
- Joint Student Supervision (combined with STSMs)
- Action workshops and training school (sampling, HTS techniques, Bioinformatics)
- 2-4 WG reports
Year 4 Milestones
- 2 MC/CG and WG meetings (shortly before or after MC meeting)
- at least 4-6 STSMs
- Joint Student Supervision (combined with STSMs)
- Action workshop and training schools
- Large international Action conference
- Presentation of ‘Cookbook’
- Final evaluation report
3.1.2 Risk and Contingency Plan
Several risks to the success of the Action have been identified that could impact on the outome. However, remediation methods are outlined in Table
Overview over potential risks and impacts on project parts and anticipated remediation methods.
|
Title of the risk |
Impact on project |
Remediation method |
|
Limited involvement of researchers in the Action |
Too few researchers involved in the Working Groups, making harmonisation and standardisation difficult. |
Already now over 30 researchers from over 20 countries have expressed their interested in this Action, making it unlikely to fail. |
|
Standard barcodes too long for working with degraded eDNA |
Impossibility to rely on the standard reference database. |
Design of new shorter barcodes, and build of a new reference database. |
|
Too many alternative technical possibilities for producing the DNA data |
Difficulty to agree on a standard. |
Set up of comparative experiments by the different groups, and organisation of a joint meeting to decide about the standard methodology (decision based on quantitative arguments). |
|
Limited engagement by stakeholders |
The topic will be restricted to academia and not find entry into EU-wide ecological assessments. |
By now there are established contacts to many dozens of stakeholders from EU- to local scale. Thus, only sporatic drop-outs are expected. |
|
Funding for national barcoding projects of EU-13 countries limited |
Reference barcode libraries will remain incomplete for several countries and taxa. |
National barcoding projects have accepted to cover sequencing (up to 10.000) from ITCs. |
3.2. Management Structures and Procedures
This COST Action is proposed by researchers from 14 COST member states, including 4 ITCs, 3 IPCs and 1 NNC. Participation from all EU member states as well as of further IPCs and NNCs is expected. The organization of the network is shown in Fig.
Overview of the structure of DNAqua-Net and interplay between the five Working Groups and the Management Committee.
Management Committee (MC): The MC of DNAqua-Net will coordinate and communicate the roadmap of the 5 WGs and ensure that milestones are reached. Delegates are expected to play a key role in the work, participate in meetings, connect to researchers as well as stakeholders in their respective countries. Furthermore, the MC will ensure that COST policies are followed, and specifically encourage active involvement of Early Career researchers. Therefore, the MC shall consist of senior scientists as well as delegates of Early Career researchers. The MC is led by the Chair and Vice-Chair. It will contain at least members of all participating countries and meet twice per year.
MC Chair: The MC chair will be the reference point for the Action, chair the annual conference/meetings (together with the MC), be responsible for the preparation of all scientific reports and the final report. The MC chair will be elected during the first meeting of the MC.
MC Vice-Chair: The MC Vice-Chair is also elected through the MC and should represent a different research field than the MC Chair. The MC Vice Chair will primarily focus on practical issues (organisation of the Action) and represent the MC in relation to the “external world”.
WG leaders: Each WG will have two leaders from different countries/research backgrounds. Junior researchers shall be actively promoted to take a lead in the WG. The WG leaders will coordinate the WG networking and capacity building activities, stimulate STSMs and contacts with other WGs. The WG leaders are in charge of further subdividing the working groups into sub-groups, coordinate the progress of these and preparing the WG output for the MC reports.
Training, Dissemination, and Liaison Manager (TDL): Will support the MC and WG leaders in the management and organisation of meetings. Organize training schools and STSMs.
Technical Manager (TM): In charge of technical preparations for the meetings and host of the website, handling contact requests.
Core Group (CG): MC Chair and Vice-Chair, WG leaders, TDL and TM. The CG serves as the coordinating body and monitors the Action’s performance. Additionally, the CG will have a proactive strategy with the aim of boosting the creation of consortia for the preparation of project proposals both at a national and international (e.g. EU programs) level.
3.3. Network as a whole
The core group of this Action proposal is formed by world-leading researchers in the field of environmental genomics, bioinformatics, and DNA-barcoding. This core group will form the scientific backbone of DNAqua-Net and represents the scientific critical mass, geographical extent, and expertise ranging from DNA barcoding to high-throughput eDNA metabarcoding, all needed to achieve the objectives set forth in the proposal. While most included institutions already have HTS facilities or the respective expertise, their role as national connection nodes will be further solidified through this Action. As the goal of DNAqua-Net is to advance ecological assessment beyond the traditionally used form, several applicants from COST member states but also NNCs with long expertise in the traditional monitoring of aquatic ecosystems are included. These partners will add to the scientific critical mass and assure the applied relevance of project goals through their long standing bioassessment expertise and good connections to water managers and other key stakeholders including policy makers. Also in NNCs and many of the ITCs there is a unique taxonomic expertise that will be of great value in order to meet the challenge of completing DNA barcode data bases
Thus, with over 20 proposers spanning all major ecoregions of Europe, this trans-disciplinary Action network connects scientific excellence with the relevant applied bioassessment knowledge to develop standards for novel eco-genomic monitoring of aquatic ecosystems. In addition, extensive collaborations with European institutions and related ongoing projects (EWA, EIP Water, as well as DNA-barcoding initiatives) will ensure that the deliverables of this Action will find entry into real-world monitoring and relevant legislation.
Through strategic partnerships with researchers and institutions from NNCs and IPCs, DNAqua-Net will further ensure that relevant trends and strategies being developed in other non-European countries are quickly detected. Further, these connections allow for fast dissemination of research findings across NNC and IPC countries through the respective national experts. Although not all European countries are presently included in the Network of Proposers, we strive to actively include many more countries after the inauguration of the Action through existing partnerships with EWA and EIP Water. Specifically, we will establish partnership with the Centre for Biodiversity Genomics, University of Guelph, to ensure that all new barcoding data are made available on a global scale through the International Barcode of Life project.
The proposers of this Action already come from various European countries including also several states of the new EU-13 countries. While the application of genomic techniques for ecological assessment is still more predominant in the old EU-states, this Action will actively invest in the training of researchers specifically from the new member states to close this gap. To achieve this goal, funds for the participation of researchers from new EU countries will be allocated through this Action.
Funding program
European Cooperation in Science & Technology program (EU COST)
Grant title
Developing new genetic tools for bioassessment of aquatic ecosystems in Europe (DNAqua-Net)
Hosting institution
University of Duisburg-Essen, Aquatic Ecosystem Research, Universitaetsstr. 5, D-45141 Essen, Germany
References
-
Three hundred ways to assess Europe's surface waters: An almost complete overview of biological methods to implement the Water Framework Directive.Ecological Indicators18:31‑41. https://doi.org/10.1016/j.ecolind.2011.10.009
-
Genomics in marine monitoring: New opportunities for assessing marine health status.Marine Pollution Bulletin74(1):19‑31. https://doi.org/10.1016/j.marpolbul.2013.05.042
-
Environmental monitoring using next generation sequencing: rapid identification of macroinvertebrate bioindicator species.Frontiers in Zoology10(1):45. https://doi.org/10.1186/1742-9994-10-45
-
From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments.Environmental research111(7):978‑88. https://doi.org/10.1016/j.envres.2011.02.001
-
Transport Distance of Invertebrate Environmental DNA in a Natural River.PLoS ONE9(2):e88786. https://doi.org/10.1371/journal.pone.0088786
-
Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences270(1512):313‑321. https://doi.org/10.1098/rspb.2002.2218
-
Environmental DNA - a review of the possible applications for the detection of (invasive) species.Stichting RAVON Report 2013104https://doi.org/10.13140/RG.2.1.4002.1208
-
The European Water Framework Directive at the age of 10: a critical review of the achievements with recommendations for the future.The Science of the total environment408(19):4007‑19. https://doi.org/10.1016/j.scitotenv.2010.05.031
-
Absolute quantification by droplet digital PCR versus analog real-time PCR.Nature methods10(10):1003‑5. https://doi.org/10.1038/nmeth.2633
-
Is DNA Barcoding Actually Cheaper and Faster than Traditional Morphological Methods: Results from a Survey of Freshwater Bioassessment Efforts in the United States?PLoS ONE9(4):e95525. https://doi.org/10.1371/journal.pone.0095525
-
Ten years of next-generation sequencing technology.Trends in Genetics30(9):418‑426. https://doi.org/10.1016/j.tig.2014.07.001
-
Eukaryotic plankton diversity in the sunlit ocean.Science348(6237):1261605‑1261605. https://doi.org/10.1126/science.1261605
-
Environmental Monitoring: Inferring the Diatom Index from Next-Generation Sequencing Data.Environmental science & technology49(13):7597‑605. https://doi.org/10.1021/es506158m